Figure 1.
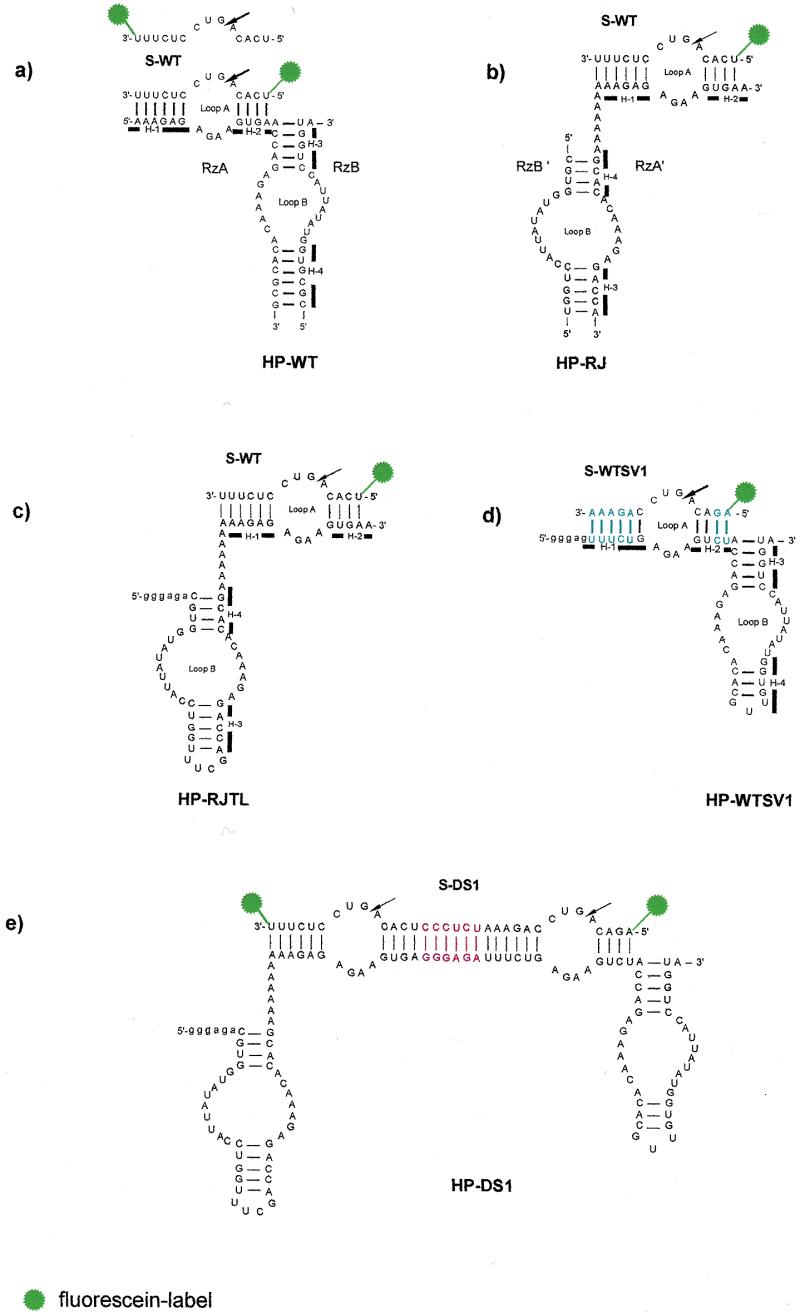
Secondary structures of the studied ribozymes. The four helices (H-1–H-4) in HP-WT, HP-RJ, HP-RJTL and HP-WTSV1 are indicated by bars. The arrows denote the sites of cleavage. Vector nucleotides resulting from in vitro transcription of HP-RJTL, HP-WTSV1 and HP-DS1 with T7 RNA polymerase are indicated in lower case. (a) Conventional hairpin ribozyme HP-WT with wild-type sequence. The fluorescently 5′- and 3′-labelled substrate S-WT is shown. (b) Reverse-joined hairpin ribozyme HP-RJ derived from HP-WT by dissecting the loop A and loop B domains at the hinge and rejoining them via an A6 linker in reverse order. (c) Reverse-joined ribozyme HP-RJTL derived from HP-RJ with helix 3 capped by the UUCG tetraloop. (d) Conventional hairpin ribozyme with sequence variation HP-WTSV1. Base pairs which are reversed in comparison to HP-WT are highlighted in blue. (e) Twin ribozyme HP-DS1 derived from combination of the monoribozymes HP-RJTL and HP-WTSV1. The sequence introduced as spacer between both ribozyme units is highlighted in red.
