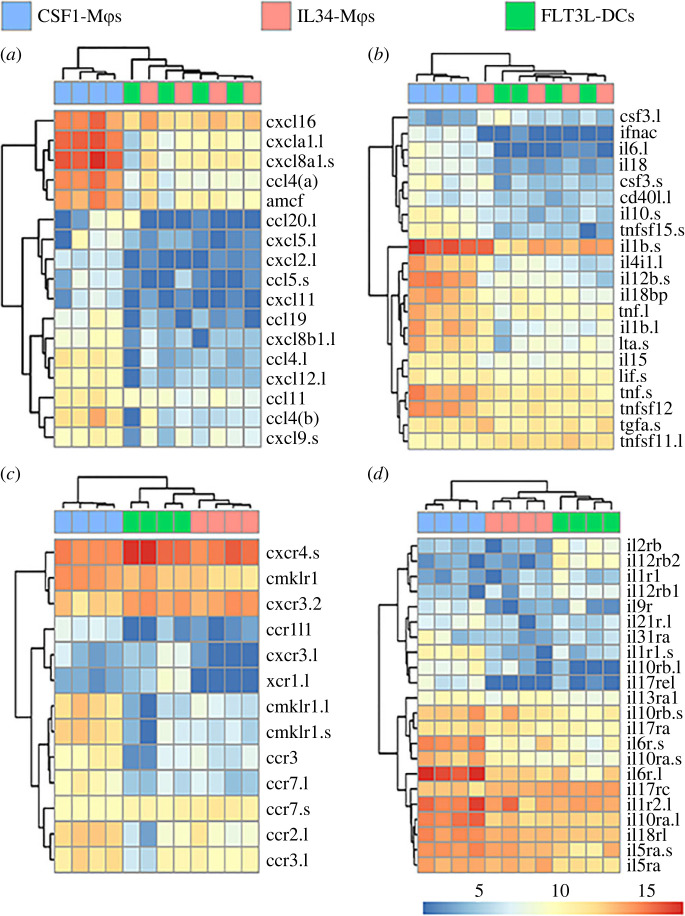
Figure 3.
Differentially expressed chemokine and cytokine ligand and receptor genes between X. laevis CSF1-Mϕs, IL34-Mϕs and FLT3L-DCs. Heatmap plots were curated to depict differentially expressed immune genes: (a) hallmark chemokine genes, (b) cytokine genes, (c) chemokine receptor genes, and (d) cytokine receptor genes were compared between all three cell types (n = 4/cell type). All plotted genes were selected from the statistically significant (as defined by padj value of less than 0.05 and absolute log2 fold change > [1]) differentially expressed genes across all tested comparisons. For each comparison, individual samples are shown in columns and immune genes in rows with colour representation of relative expression, as indicated by the colour bar. Any gene names that appear multiple times within respective heat plots were detected as distinct based on model identities, being transcripts from distinct loci and/or being transcript from short and long arm of X. laevis chromosomes, as indicated by ‘.s’ or ‘.l' suffixes, respectively. All heatmaps were visualized using ‘pheatmap' package in R (4.0.2 version). (Online version in colour.)