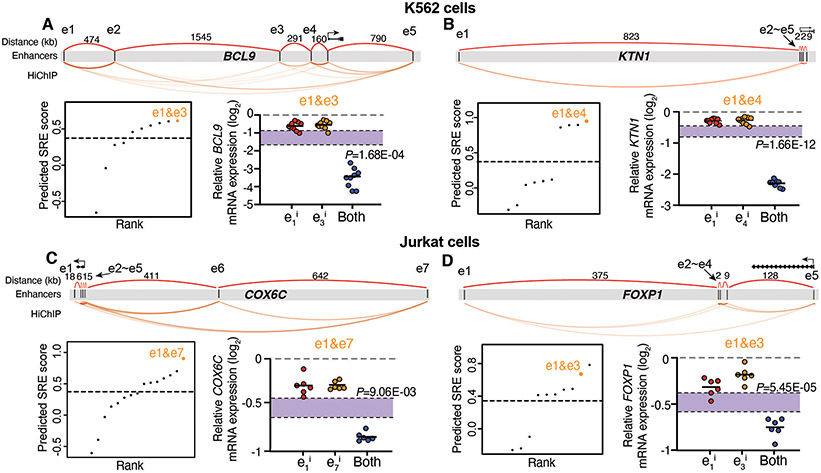
Fig. 3. Experimental validation of predicted SREs at other genomic loci in different cell types.
A-D, Prediction and validation of SREs at BCL9 (A) and KTN1 loci (B) in K562 cells, and COX6C (C) and FOXP1 loci (D) in Jurkat cells. Top: Diagram showing multiple enhancers spanning an ultralong distance at each genomic locus. Bottom left: Rank of predicted SREs using the model. Dashed line represents the empirical threshold from the MYC locus. Orange dots indicate the validated SREs. Bottom right: qRT-PCR of mRNA expression for each gene when perturbing the predicted SREs. Data are represented as individual biological replicates (dots) and the mean value (black bar). The purple area indicates the expected additive effect by plotting mean ± one standard derivation. P values are calculated by t-test.