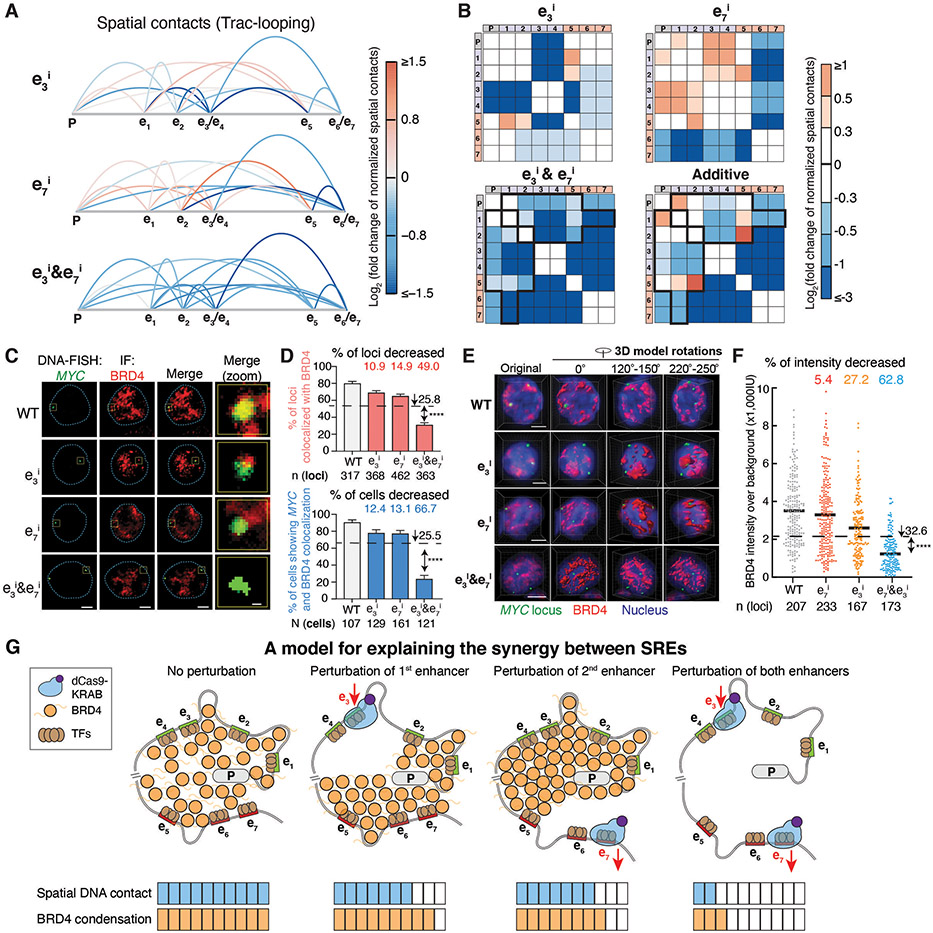
Fig. 4. Perturbation of SREs leads to synergistic reduction of spatial contacts and BRD4 condensation at the genomic locus.
A-B, Spatial contacts between the promoter and enhancers measured by Trac-looping for the MYC locus upon perturbation of e3, e7, and e3&e7. Colors represent the log2 fold change of spatial contacts normalized to the wildtype cells. Black boxes in (B) indicate synergistically decreased (more than additive) spatial contacts of e3&e7 pair perturbation.
C-F, DNA-FISH colocalization between BRD4 and the MYC locus of representative K562 cells for 2D (C-D) and 3D image analysis (E-F) upon perturbation of e3, e7, and e3&e7. In (C&E), red, BRD4 immunofluorescence (IF) staining; green, DNA-FISH at the MYC locus; blue dashed line, nuclear periphery determined by DAPI staining (not shown); scale bars, 5μm. The rightmost column in (C) shows insets in the yellow boxes. Scale bars, 500 nm. Quantification of BRD4 and the MYC locus colocalization are shown for 2D (D) and 3D image analysis (F). In (D), percentage of loci with colocalization is shown on the top and percentage of cells (≥ 2 colocalization loci) is shown on the bottom; data is represented as mean ± standard error of the mean. In (F), each dot represents an individual locus. n = total loci, N = total cells. **** P < 0.0001 in Fisher’s exact test (D) or t-test (F) versus the expected additive effect (dashed line).
G, A model to explain the synergy between SREs.