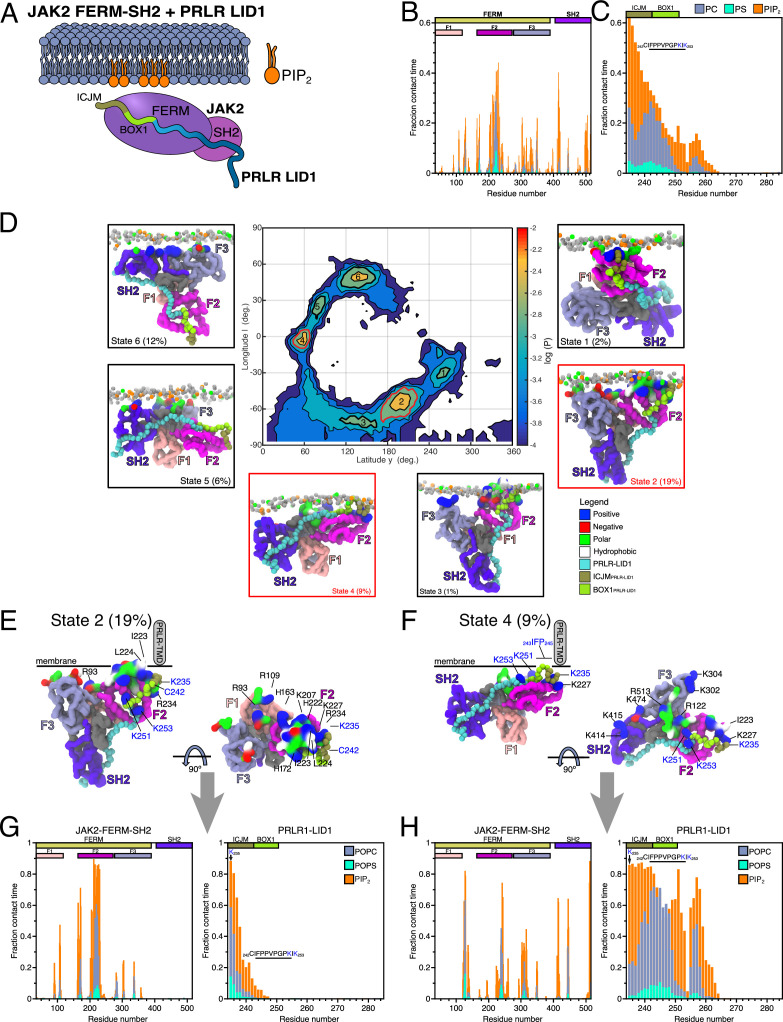
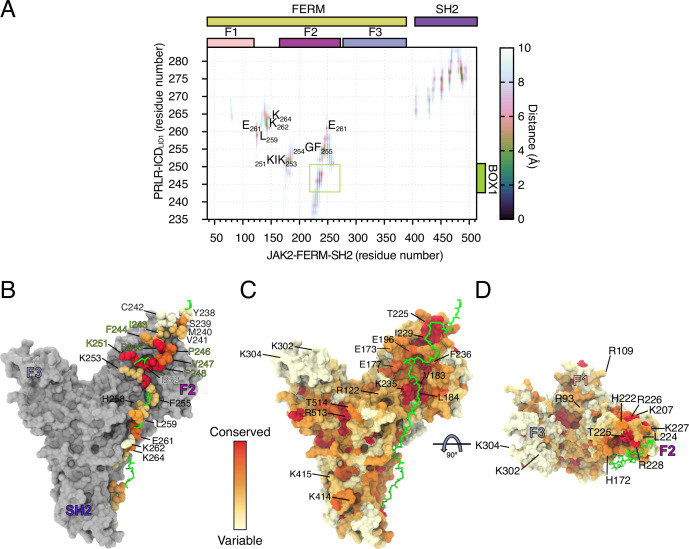
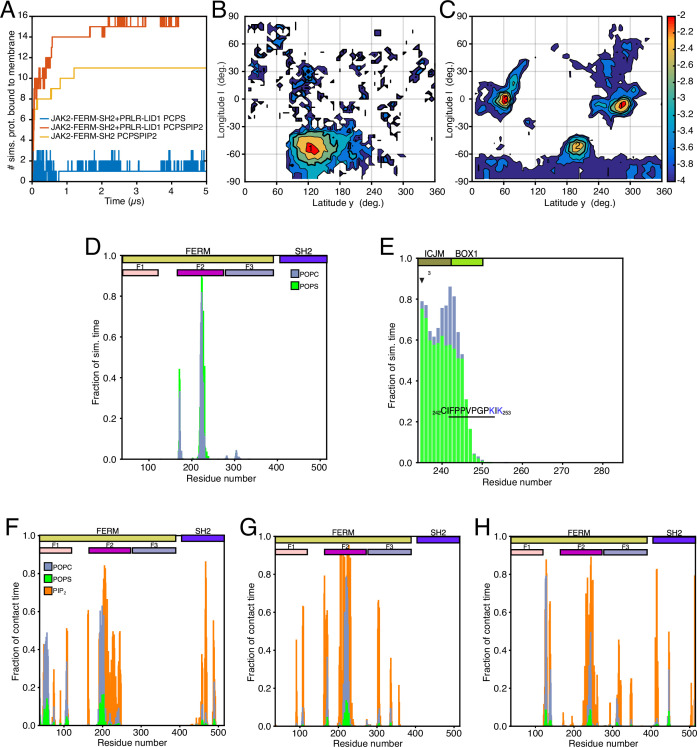
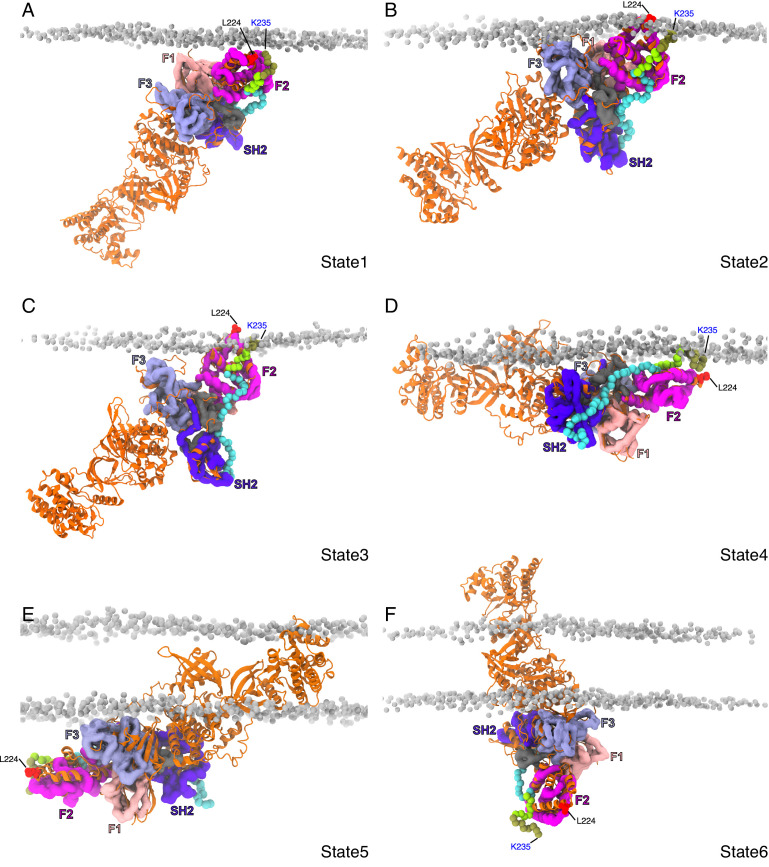
Figure 4. Protein–lipid interactions of the JAK2-FERM-SH2 PRLR-ICDLID1 complex obtained from coarse-grained (CG)-molecular dynamic (MD) simulations.
(A) Schematic representation of the simulated system. Combined (B) JAK2-FERM-SH2-lipid and (C) PRLR-ICDLID1-lipid contact frequency histograms for the 16 CG simulations of the JAK2-FERM-SH2+PRLR-ICDLID1+POPC:POPS:PI(4,5)P2 system. (D) Distribution of the orientations adopted by the JAK2-FERM-SH2+PRLR-ICDLID1 complex when bound to lipids taken from the 16 simulations with POPC:POPS:PI(4,5)P2 in the lower-leaflet. The snapshots surrounding the map correspond to representative conformations of the highlighted states also indicating the fraction total bound time for which each state was observed. Representative conformations of (E) State 2 and (F) State 4. The gray cylinder depicts the position where PRLR-transmembrane domain (TMD) should be located. Representative protein–lipid contact histograms for (G) State 2 and (H) State 4 colored as in panels B and C. POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; POPS, 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine.