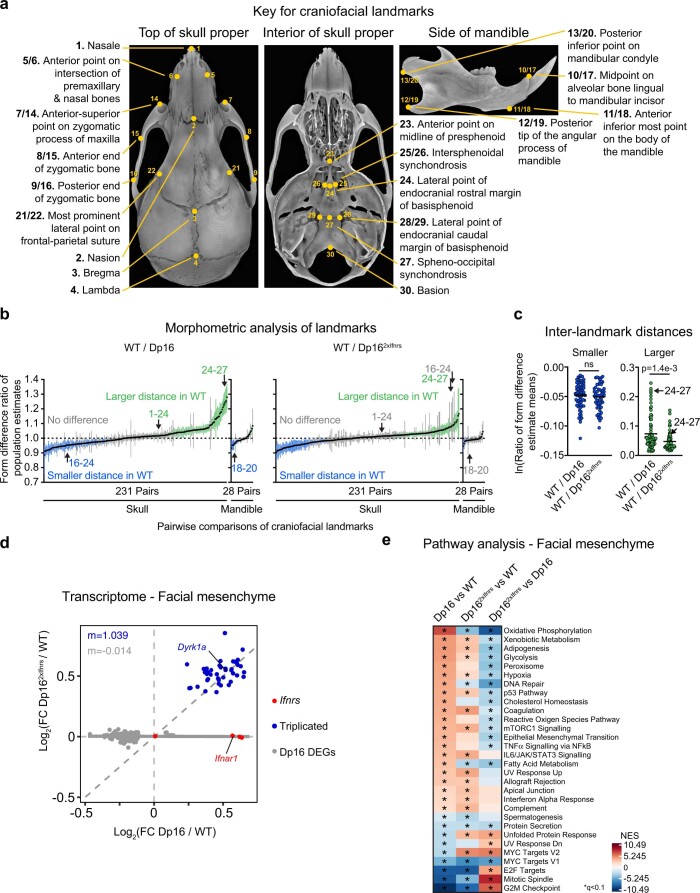
Extended Data Fig. 6. Triplication of the Ifnr locus exacerbates craniofacial morphometric differences in a mouse model of Down syndrome.
a, Representative micro-computed tomography (μCT) reconstructions of a wild-type (WT) mouse skull. Landmarks are indicated on dorsal views of the outer skull proper (left) and interior cranial base (center) as well as on a lateral view of the hemi-mandible (right). b, Form difference ratio of inter-landmark distances for the skull proper and mandible by Euclidean Distance Matrix Analysis for cohorts of Dp16 and Dp162xIfnrs mice compared to WT controls, followed by bootstrapping 10,000x. The precise number of animals per group prior to bootstrapping are WT (n = 7, 2 male, 5 female), Dp16 (n = 7, 4 male, 3 female), and Dp162xIfnrs (n = 6, 4 male, 2 female). Data are represented as mean population estimates (black dots) with 95% confidence intervals (lines) colored according to differences versus WT where green represents larger inter-landmark distance, blue represents smaller inter-landmark distance, and gray represents confidence intervals that intersect 1 (that is, no difference). c, Distributions of form difference ratios (natural log (ln)-transformed) for all mean population estimates of inter-landmark distances on the skull proper and mandible shown in panel (b) that were smaller (blue) or larger (green) in WT versus Dp16 or versus Dp162xIfnrs (smaller, blue, or larger, green). WT (n = 7, 2 male, 5 female), Dp16 (n = 7, 4 male, 3 female), and Dp162xIfnrs (n = 6, 4 male, 2 female). p-values determined by two-sided Mann-Whitney test with significance set at p < 0.05. d, Scatter plots comparing mRNA fold-changes for Dp16 differentially expressed genes (DEGs) in facial mesenchyme tissue from E10.5 mice for Dp16/WT and Dp162xIfnrs/WT, with Ifnrs highlighted in red, Dp16 triplicated genes in blue, non-triplicated Dp16 DEGs in gray, and slope (m) colored accordingly; solid gray lines represent linear fits for the non-triplicated Dp16 DEGs. WT (n = 3, 2 male, 1 female), Dp16 (n = 3, 2 male, 1 female), Dp162xIfnrs (n = 3, 2 male, 1 female). e, Heatmap displaying Normalized Enrichment Scores (NES) from Gene Set Enrichment Analysis (GSEA) of transcriptome changes in E10.5 facial mesenchyme for indicated comparisons, sorted by the for Dp16 versus WT comparison; asterisks indicate q < 0.1 by GSEA after Benjamini-Hochberg correction.