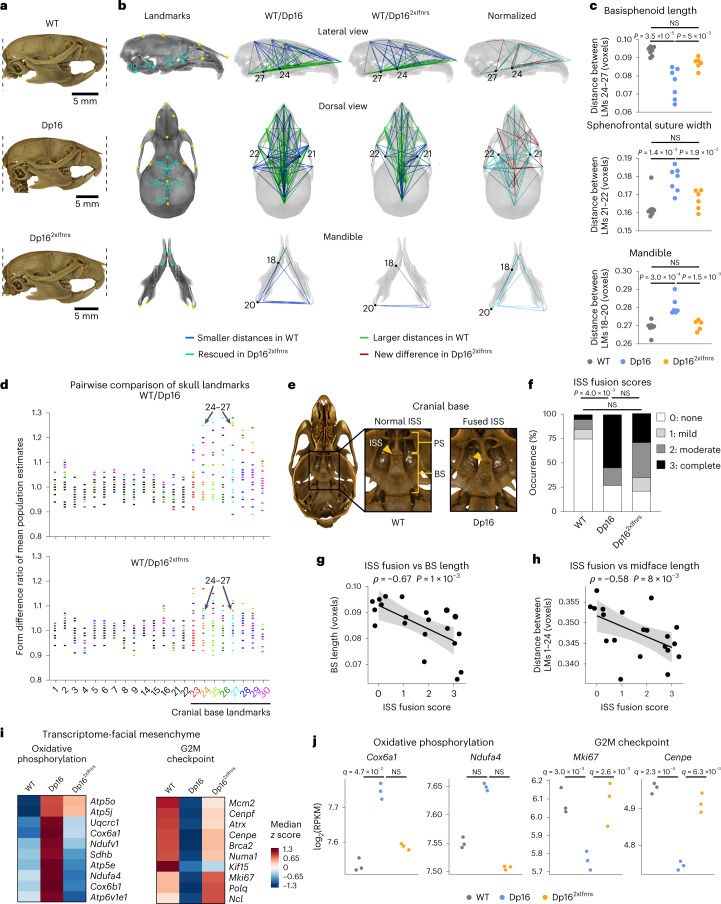
Fig. 6. Triplication of the Ifnr locus exacerbates craniofacial anomalies in a mouse model of DS.
a, Representative lateral views of μCT reconstructions of skulls from WT, Dp16 and Dp162xIfnrs animals, aligned and scaled based on the same 3D linear measurement. b, Lateral (top) and dorsal (middle) views of the outer portion of the skull proper transparently overlaid on cranial base interior view from a WT mouse, with landmarks on the skull (top and middle) and mandible (bottom) in yellow, and interior landmarks on the cranial base in turquoise. Smaller and larger interlandmark distances (blue and green, respectively) in WT relative to Dp16 or Dp162xIfnrs calculated by EDMA. Number of animals—WT (n = 7, 2 male and 5 female), Dp16 (n = 7, 4 male and 3 female) and Dp162xIfnrs (n = 6, 4 male and 2 female), followed by bootstrapping 10,000 times. Turquoise—distances different in WT relative to Dp16 rescued in Dp162xIfnrs. Red—distances with no difference in WT relative to Dp16 but different in Dp162xIfnrs. c, Example interlandmark distances before bootstrapping. Number of animals as in b. P values determined by one-way ANOVA with Tukey’s HSD test, with significance set at P < 0.05; horizontal dashes indicate means. d, Form difference ratios of mean population estimates for distances on the skull after bootstrapping, with colors assigned to any distance that includes a cranial base landmark. e, Images of cranial base interior views for WT and Dp16 displaying normal or completely fused ISS. f, Frequency of complete ISS fusion (black) compared between cohorts by pairwise two-sided Fisher’s exact test with significance set at P < 0.05. Number of animals—WT (n = 20, 12 male and 8 female), Dp16 (n = 11, 6 male and 5 female) and Dp162xIfnrs (n = 14, 7 male and 7 female). g,h, Scatter plots comparing ISS fusion scores to BS length (g) or midface length (h) for each skull where total n = 20 (numbers per genotype and sex as in b), showing Spearman ρ and P values (permutation test). Black lines represent linear fits with 95% CIs shaded in gray. i, Heatmap displaying median RPKM expression of z scores for example genes dysregulated in E10.5 facial mesenchyme. Number of animals—WT (n = 3, 2 male and 1 female), Dp16 (n = 3, 2 male and 1 female) and Dp162xIfnrs (n = 3, 2 male and 1 female). j, Sina plots displaying expression levels for example genes; q values determined by DESeq2 with Benjamini–Hochberg correction, with significance set at q < 0.1. PS, presphenoid bone.