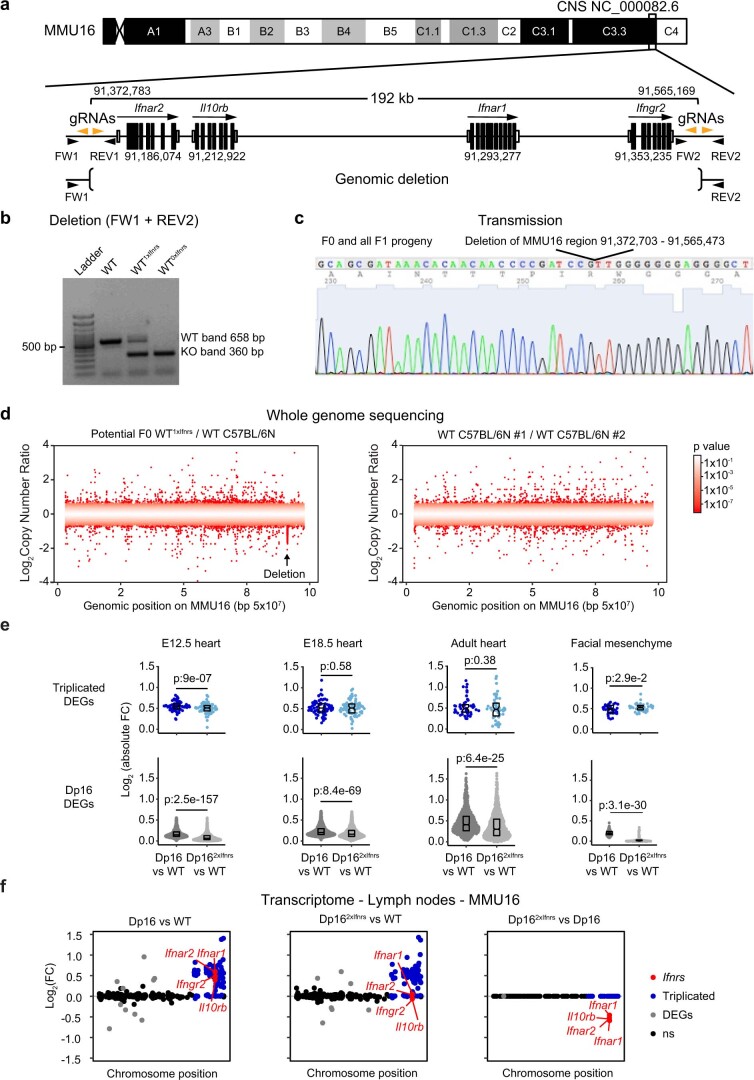
Extended Data Fig. 2. Mouse model to determine if triplication of the Ifnr locus on mouse chromosome 16 is necessary for Down syndrome phenotypes.
a, Schematic of the mouse interferon receptor (Ifnr) gene locus and CRIPSR/Cas9 guide RNAs (gRNAs, orange arrowheads) used for deletion of 192 kb genomic locus on chromosome 16 (MMU16). Black arrows indicate forward (FW) and reverse (REV) primers. Location of genes based on GRCm38 reference genome is indicated on an ideogram of MMU16 cytogenetic regions colored according to Giemsa banding. b, Representative PCR from DNA of wild-type (WT) mouse, and mice heterozygous or homozygous for the expected knock-out (KO) deletion (WT1xIfnrs or WT0xIfnrs, respectively). Gel image represents example of genotyping PCR used to characterize >50 pups derived from modified zygotes. This approach screened 5113 different descendants to date from a single heterozygous founder (F0). c, Representative Sanger sequencing of the single modified allele transmitted from a F0 WT1xIfnrs male to the first generation of progeny (F1) after inter-crossing with a WT female. d, Whole genome sequencing followed by copy number variant analysis of the F0 WT1xIfnrs with site of deletion on MMU16 denoted by arrow (left) that is absent when two non-related C57BL/6 N WT mice are compared (right). Significance was determined by CNV-seq (*p < 0.1). e, Transcriptome analysis of hearts at embryonic day (E)12.5 - WT (n = 6, 2 male, 4 female), Dp16 (n = 6, 3 male, 3 female), Dp162xIfnrs (n = 6, 5 male, 1 female), E18.5 - WT (n = 6, 4 male, 2 female), Dp16 (n = 6, 3 male, 3 female), Dp162xIfnrs (n = 6, 3 male, 3 female), adult - WT (n = 5, 2 male, 3 female), Dp16 (n = 6, 3 male, 3 female), Dp162xIfnrs (n = 5, 3 male, 2 female), and facial mesenchyme at E10.5 - WT (n = 3, 2 male, 1 female), Dp16 (n = 3, 2 male, 1 female), Dp162xIfnrs (n = 3, 2 male, 1 female). Sina plots of mRNAs encoded on MMU16 triplicated in Dp16 excluding the four Ifnrs (top), and other differentially expressed genes (DEGs) across the genome for Dp16 versus WT (bottom). p-values calculated by two-sided paired Wilcoxon rank test. Data are presented as modified sina plots where boxes represent interquartile ranges and medians, and notches approximate 95% confidence intervals. f, Manhattan plots of mRNAs encoded on MMU16 differentially expressed by genotype in the mesenteric lymph nodes and colored as indicated. *q < 0.1 determined by DESeq2 with Benjamini-Hochberg correction.