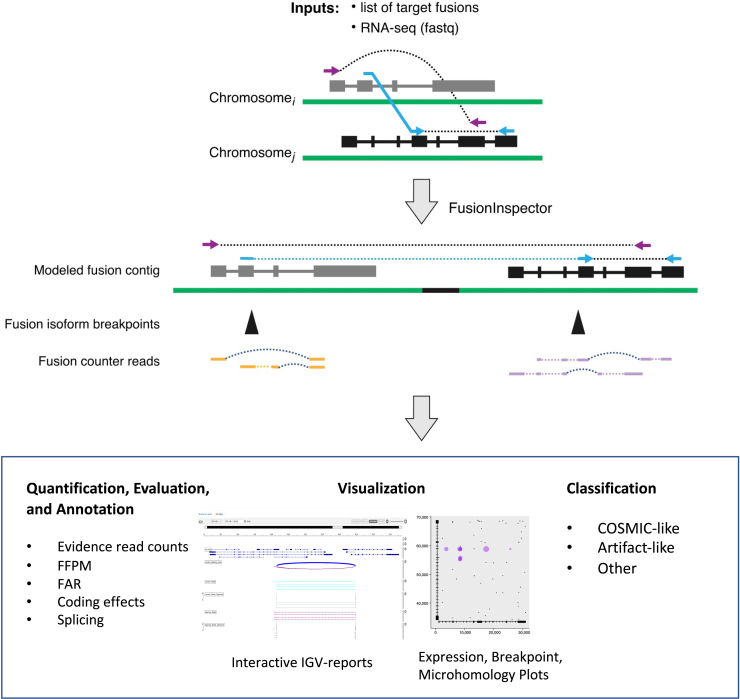
Figure 1.
FusionInspector overview
Top: lists of fusion candidates derived from predictions of one or multiple fusion detection methods, or from a screening panel, are provided to FusionInspector as input along with RNA-seq in fastq format. For each candidate fusion gene, fusion contigs are generated by fusing the full-length gene candidates as collinear on a single contig. Intronic regions are by default each shrunk to 1 kb. RNA-seq reads are then aligned to a reference consisting of the entire genome supplemented with fusion contigs. Fusion-derived reads that would normally align discordantly as chimeric alignments in the reference genome (top example) instead align concordantly in the fusion contig context (bottom example).
Middle: FusionInspector identifies split-read alignments (light blue) and spanning pairs (purple) supporting the gene fusion in addition to read alignments that overlap the breakpoints and instead support the unfused fusion partners (fusion counter-reads).
Bottom: for those fusions where FusionInspector captures RNA-seq read support (“in silico validation”), it reports fusion sequence and expression characteristics, interactive visualizations (Figure S1B), and predicted classifications as COSMIC-like, potential artifact, or other category (STAR Methods).