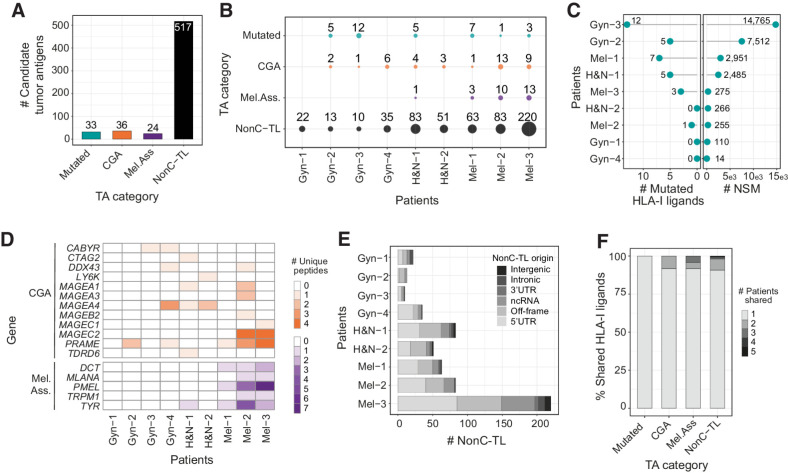
Figure 2.
Candidate tumor antigens presented on HLA-I from patient-derived TCL identified through proteogenomics. A, Total number of unique peptide sequences derived from candidate tumor antigens (TA) by category. Data from all patients were pooled together. Only unique peptide sequences were considered. B, The number and category of TA are displayed for each TCL. C, The number of mutated peptides eluted from HLA-I (left) and number of NSM identified by WES (right) are displayed for each patient. D, Heat map displaying the number of epitopes derived from specific CGA or melanoma-associated antigens per patient. E, Number of nonC-TL originated from each ORF category per patient. F, Percentage of candidate TA uniquely identified in one patient or shared by TA category. The FDR threshold was set at 0.02 for mutations and 0.01 for all other categories.