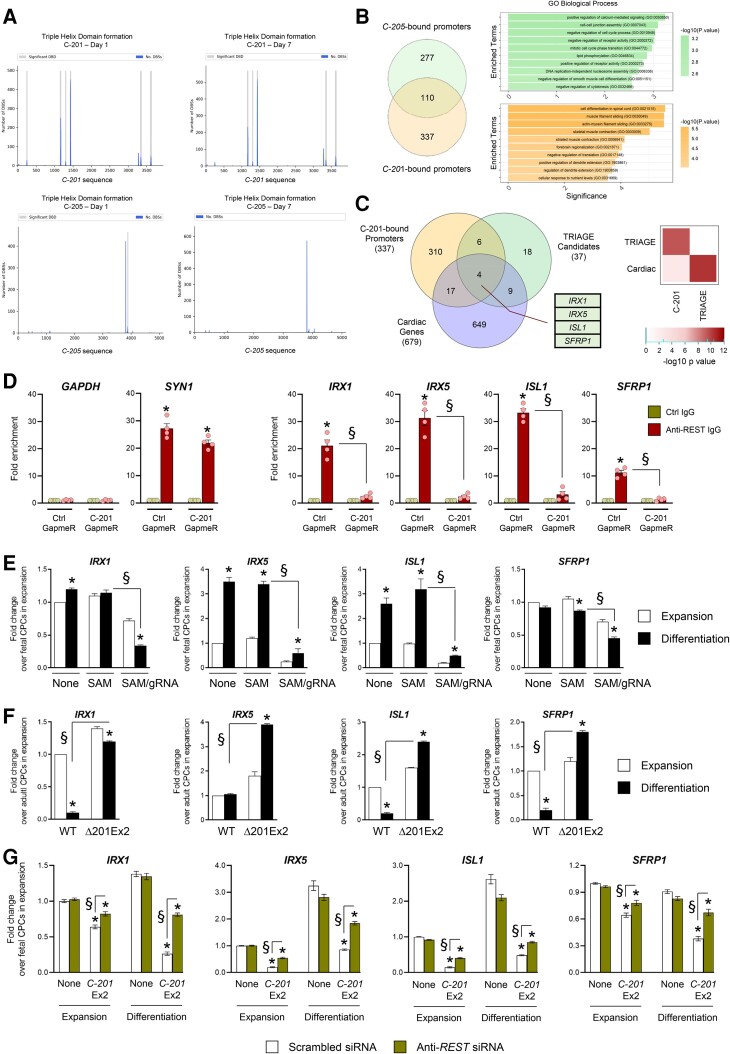
Figure 5.
Identification of IRX1, IRX5, SFRP1 and ISL1 as target genes of C-201 action. (A) Significant DNA-binding domains (DBDs) identified in the mature sequence of C-201 and C-205 when analyzed against the differentially downregulated genes on Day 1 and Day 7 following induction of differentiation. Graph shows the number of DNA-binding sites (DBS) for each DBD. (b) Venn diagram illustrating the overlap of promoters predicted to form triple helices with C-201 and C-205. Functional enrichment analysis of the isoform-specific bound promoters. Graph shows the negative logarithm of the P value. (C) Venn diagram illustrating the identification of IRX1, IRX5, ISL1, and SFRP1 as common to the indicated lists of genes. Hypergeometric tests were performed to explore the significance of the overlap (D) REST occupancy at the promoters of IRX1, IRX5, SFRP1, and ISL1 in adult CPCs with or without GapmeR-mediated C-201 silencing as determined by ChIP-qPCR. Occupancy at the GAPDH and the SYN1 promoters was used as negative and positive controls, respectively. (E) Expression of IRX1, IRX5, ISL1, and SFRP1 in differentiating fetal cells either untransfected (None), transfected with the SAM system in the absence of gRNA (SAM) or with the SAM system with a gRNA targeting sequences upstream the CARMEN TSS (SAM/gRNA). (F) Expression of IRX1, IRX5, ISL1, and SFRP1 in differentiating adult CPCs treated with Scrambled siRNA (Scr SiRNA) or Anti-REST siRNA. (G) Expression of IRX1, IRX5, ISL1, and SFRP1 in differentiating fetal CPCs either untransfected, transfected with a lentiviral vector encoding C-201 Ex2 or a mutated C-201 Ex2 (C-201 mutEx2), treated with either a scrambled siRNA (Scr siRNA) or a siRNA directed against REST (Anti-REST siRNA). Data represent mean values ± SEM; *P < 0.05 as compared with CPCs in expansion; §P < 0.05 compared with indicated conditions (n = 3–6). ANOVA with post hoc Tukey. See also Supplementary material online, Figure S7.