Figure 3.
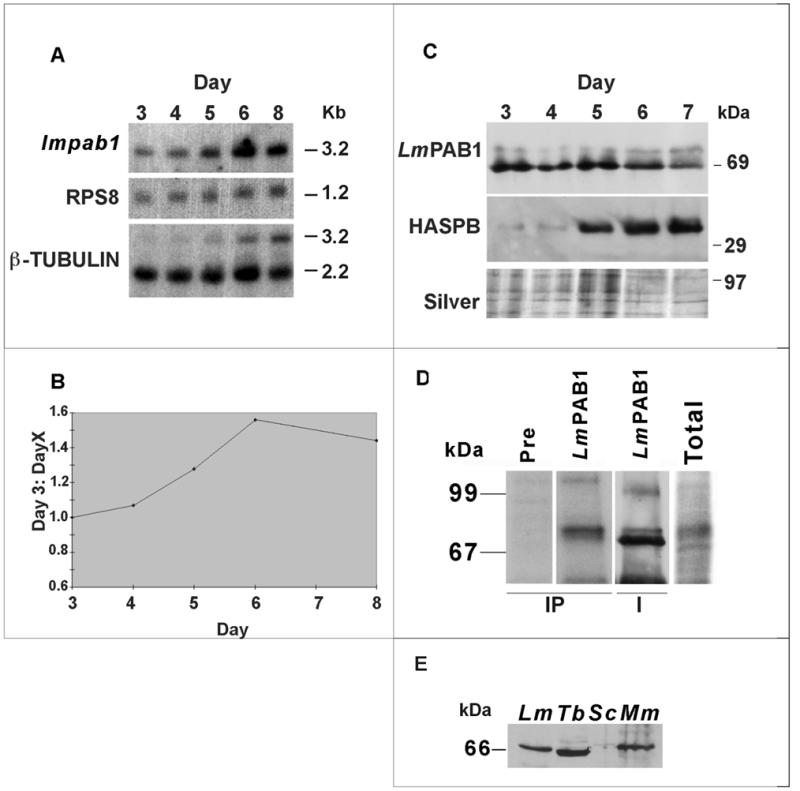
Analysis of the expression pattern of LmPAB1. (A) Total RNA, isolated from differentiating parasites over a time course (3–8 days), was size-separated, blotted and probed with the open reading frame of lmpab1. The same blot was re-probed with RPS8 (constitutively expressed, S8 ribosomal protein gene) and β-tubulin (which recognises differentially expressed transcripts of 2.2 and 3.2 kb; 45). (B) Expression of the lmpab1 transcript normalised against RPS8 expression and plotted as a ratio of day 3 (logarithmic parasite) expression levels. (C) Expression of LmPAB1 in differentiating parasites. Total proteins were extracted at daily intervals (3–7 days), separated by 10% SDS–PAGE, electroblotted and identical blots probed with abSK375 (anti-LmPAB1, top panel) and ab366 (anti-HASPB, middle panel). Silver staining was used to visualise protein loading (bottom panel). (D) In vivo phosphorylation of LmPAB1. Total lysate from 32P-radiolabelled parasites was immunoprecipitated (IP tracks) with anti-LmPAB1 or pre-immune serum, prior to analysis by 8% SDS–PAGE. The IP/LmPAB1 track was then immunoblotted against anti-LmPAB1 (I track). Relative exposure times at –70°C: total, 1 × 107 parasites labelled with 100 µCi 32P, 14 h; IP, 1 × 108 parasites labelled with 200 µCi 32P, 96 h. (E) Cross-reactivity of anti-LmPAB1with PAB1 from other species. Cell extracts from L.major (Lm, 5.0 × 106), T.brucei procyclics (Tb, 2.0 × 107), S.cerevisiae (Sc, 2.0 × 107) and mouse macrophages (Mm, 2.0 × 106) were separated by 10% SDS–PAGE and immunoblotted with anti-LmPAB1.
