Fig. 4.
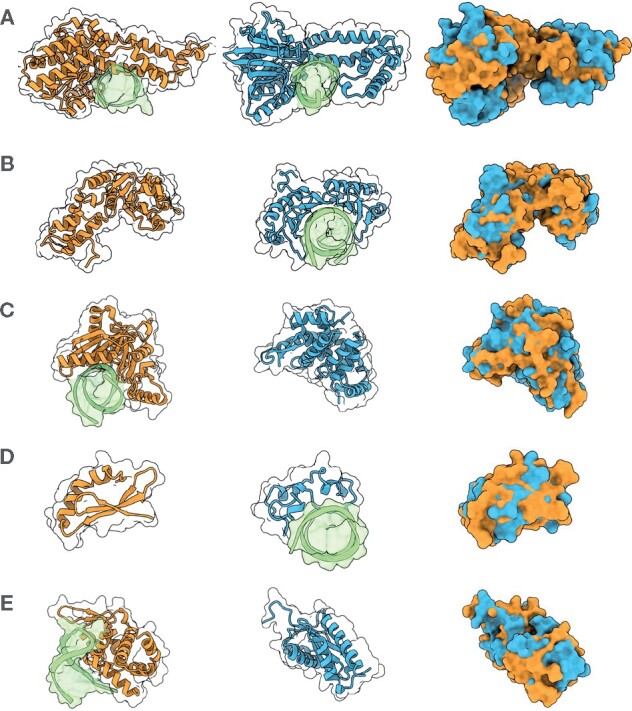
A selection of shape matches and their ZEAL-alignments in the 2S set that are all annotated with the keyword ‘DNA-binding’ (A–C) and ‘zinc-finger’ (D, E) in UniprotKB, and with TM-scores under 0.3. DNA structures in the PDB entries are shown in green. (A) Ecl18kI restriction endonuclease [2GB7 chain A] (orange) and BsoBI restriction endonuclease [1DC1 chain A] (blue). (B) Psp operon transcriptional activator [4QOS chain A] (orange) and TATA-binding protein [1VTO chain A] (blue). (C) Endonuclease V isoform X2 [6OZI chain A] (orange) and CRISPR-associated protein three HD domain [3SK9 chain A] (blue). (D) RLD2 BRX domain [6L0V chain E] (orange) and CpG-binding protein [3QMD chain A] (blue). (E) Roquin-2 [4ZLD chain A] (orange) and Avian sarcoma virus integrase [1CXQ chain A] (blue)
