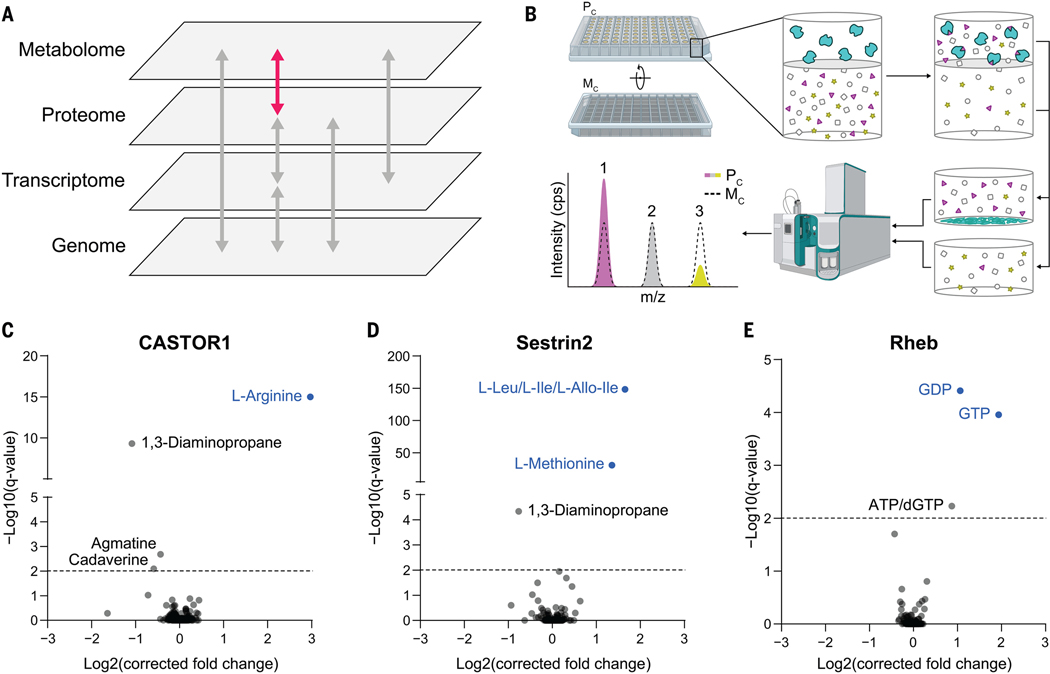
Fig. 1. MIDAS is a platform for the systematic discovery of PMIs.
(A) Biological systems are organized into domains of information (labeled gray panes). Flow of information within and between these domains is transmitted through direct interactions and underlies biological function (arrows). The MIDAS platform provides PMI discovery (pink arrow). (B) The MIDAS platform is an equilibrium dialysis tandem FIA-MS approach. (Top left and top center) Purified proteins (cyan) are loaded into the protein chamber (Pc) and defined pools of metabolites into the metabolite chamber (Mc), separated by a protein-impermeable dialysis membrane. (Top right) The system is incubated to relative equilibrium. (Bottom right and bottom center) Proteins are removed by precipitation, metabolites in the Pc and Mc are sampled, and the relative abundance of metabolites from both chambers are quantified using FIA-MS. (Bottom left) PMIs are observed as an increase (1) or decrease (3) in metabolite abundance in the Pc relative to the Mc (dotted peak). Metabolites that have equal abundance in the Pc relative to the Mc (2) are defined as noninteracting with the target protein. cps, counts per second; m/z, mass/charge ratio. (C to E) Volcano plots of MIDAS analyses of the mTORC1 regulators CASTOR1, Sestrin2, and Rheb. Significant PMIs are labeled; previously known interactions are blue. All proteins were screened by triplicate equilibrium dialysis and technical triplicate FIA-MS injections. Significant PMIs identified by MIDAS are labeled and have a Q < 0.01 (dotted line).