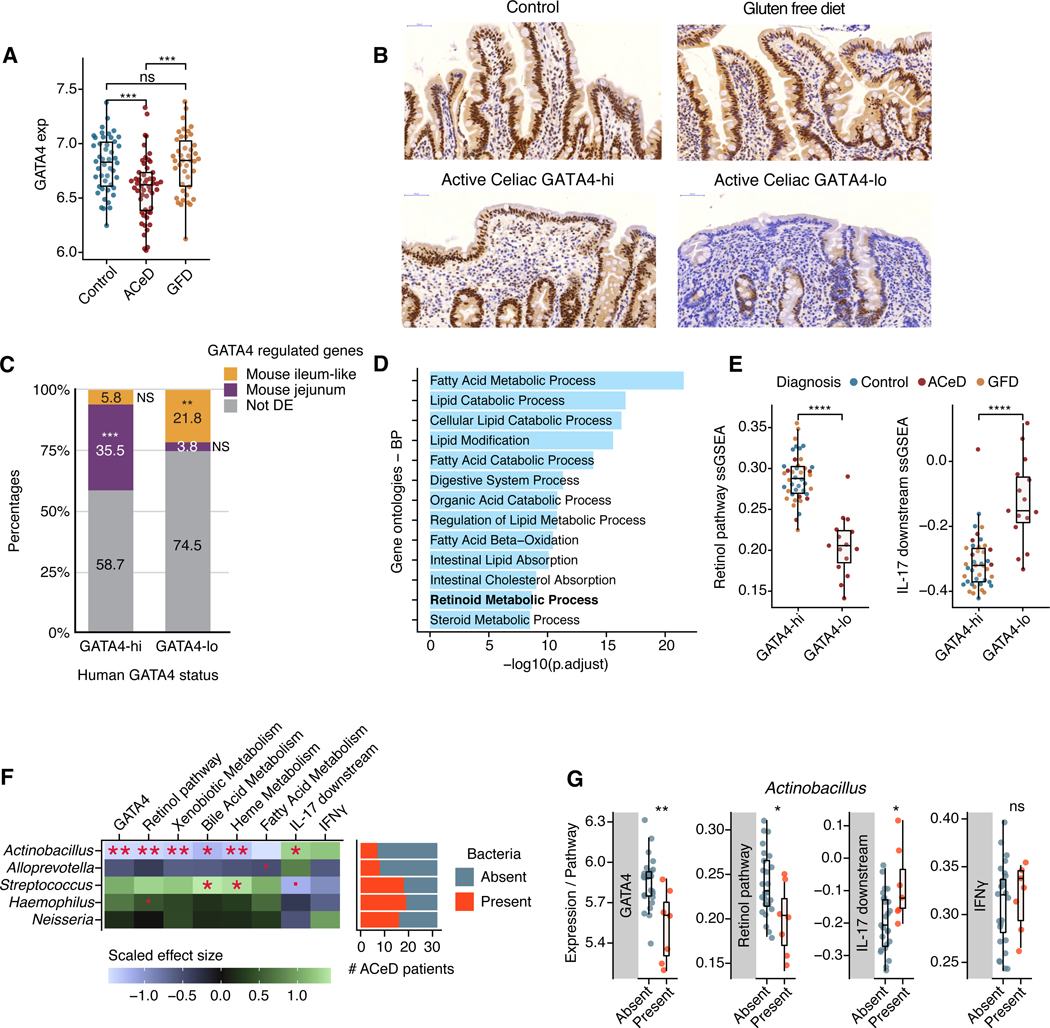
Figure 6. Loss of GATA4 expression is associated with lipid metabolic dysfunction and increased IL-17 signaling in celiac disease.
(A) Normalized GATA4 expression in duodenal biopsies from healthy controls, active celiac disease patients (ACeD), and gluten free diet celiac patients (GFD).
(B) Representative IHC staining for GATA4 in healthy control, GATA4-hi and GATA4-lo active celiac disease patients, and gluten free diet celiac patients.
(C) Bar plot shows the percentages of human-mouse homologous genes specific to GATA4-hi or GATA4-lo individuals, which are also either GATA4-regulated and specific to the jejunum (purple) or to the ileum-like tissues (yellow), or not (gray). *** P <10−51, ** P <10−6, NS not significant.
(D) Bar plot shows the most significantly enriched gene ontologies and their significance (x axis, negative log FDR-adjusted P-values) in the intersection of genes specific to GATA-hi individuals and WT mouse jejunum.
(E) Single-sample gene set enrichment analysis (ssGSEA) scores for the retinol metabolism (left) and IL-17 downstream signaling (right) pathways in GATA4-hi and GATA4-lo individuals from all patient groups.
(F) Left, heatmap displays the scaled effect size of the absence or presence of five relevant bacteria (Fig. S4I) on GATA4 expression and on the ssGSEA scores of metabolic and immune pathways. Right, bar plot shows the numbers of detectable bacteria in ACeD samples.
(G) Box plots show GATA4 expression and ssGSEA scores for the retinol metabolism, IL17 downstream signaling, and IFNγ pathways in ACeD patients, grouped by the absence or presence of Actinobacillus.
**** P<0.0001 , *** P<0.001, ** P<0.01, * P<0.05, · P <0.1, Wilcoxon rank test (A, E, G), Fisher’s exact test (C), t-test (F).