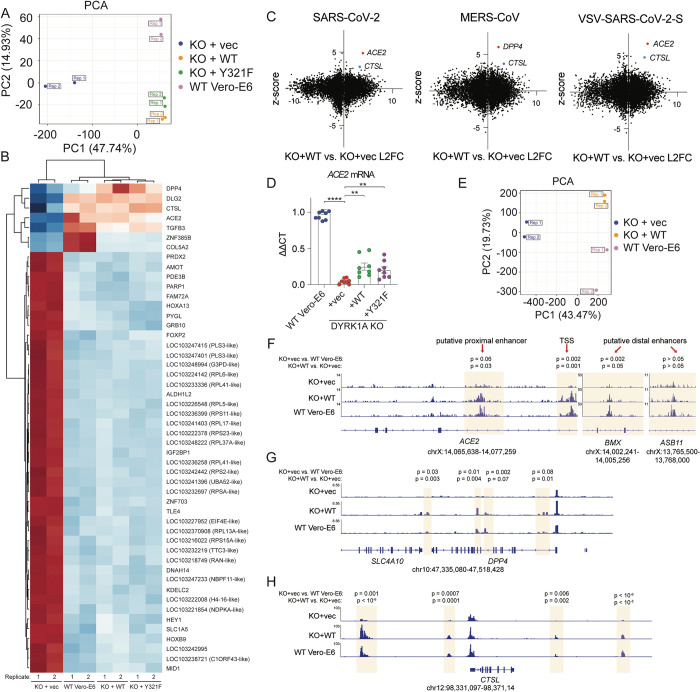
Fig 4. DYRK1A drives ACE2 and DPP4 expression by altering chromatin accessibility.
(A) Principal component analysis of RNA-Seq experiments performed in WT Vero-E6, KO+vec, KO+WT, and KO+Y321F cells. Rep refers to independent biological replicates. (B) Heatmap depicting DEGs by RNA-Seq in WT Vero-E6, KO+vec, KO+WT, and KO+Y321F cells. (C) XY plot of RNA-Seq L2FC versus CRISPR z-scores in Vero-E6 cells [23] for SARS-CoV-2 (left), MERS-CoV (middle), or VSV-SARS-CoV-2-S (right). Denoted are receptor (ACE2, DPP4) and protease (CTSL) genes as significantly up-regulated proviral genes of interest. (D) RT-qPCR for ACE2 validates RNA-Seq results and supports partial rescue of ACE2 mRNA transcripts in cells where DYRK1A-WT or DYRK1A-Y321F are reintroduced. (E) Principal component analysis of ATAC-Seq experiments performed in WT Vero-E6, KO+vec, KO+WT, and KO+Y321F cells. Rep refers to independent biological replicates. (F) ATAC-Seq gene tracks for ACE2, highlighting increased accessibility at putative enhancers and near the TSS in the presence of DYRK1A. (G) ATAC-Seq gene tracks for DPP4, showing increased chromatin accessibility in the presence of DYRK1A. (H) ATAC-Seq genome tracks for CTSL, showing increased chromatin accessibility in the presence of DYRK1A. All experiments were performed in biological duplicate (RNA-Seq/ATAC-Seq) or triplicate (RT-qPCR). Data underlying this figure can be found in S1 Data and under GEO Accession GSE213999 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE213999). ACE2, angiotensin-converting enzyme 2; CTSL, Cathepsin L; DEG, differentially expressed gene; DPP4, dipeptidyl peptidase-4; DYRK1A, Dual Specificity Tyrosine Phosphorylation Regulated Kinase 1A; KO, knockout; L2FC, log2 fold-change; MERS-CoV, Middle East Respiratory Syndrome Coronavirus; RT-qPCR, quantitative reverse transcription PCR; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; TSS, transcriptional start site; WT, wild-type.