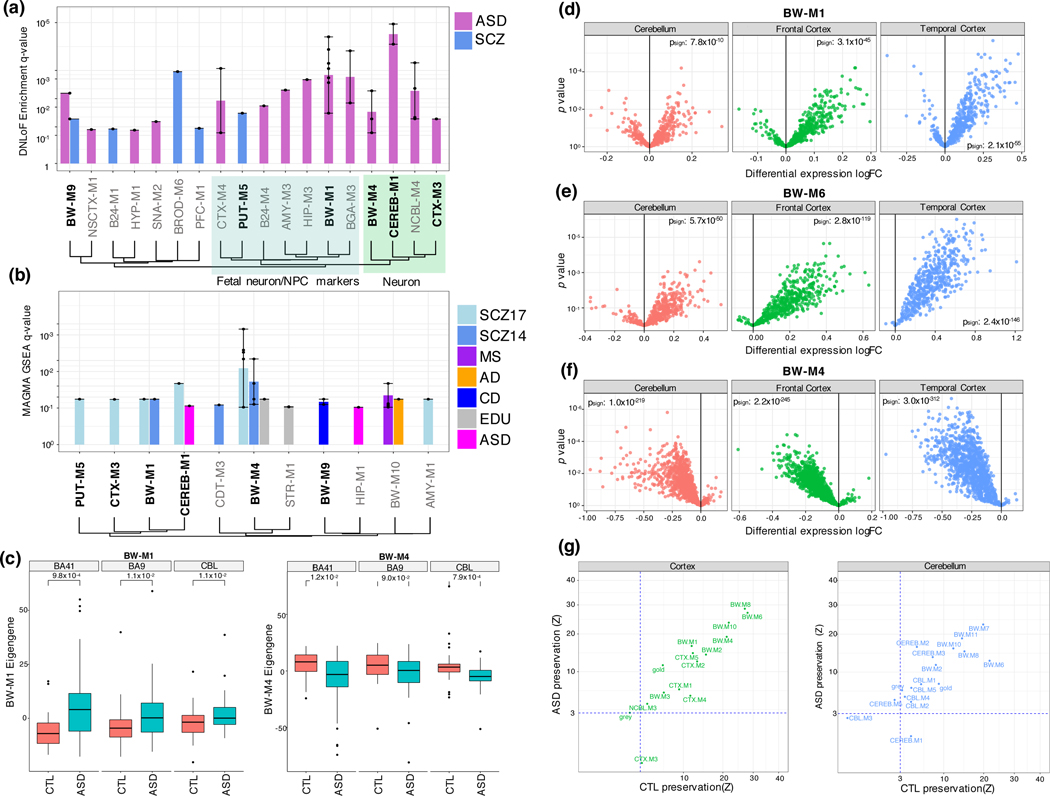
Figure 5:
Gene-level module enrichments for de novo PTVs, GWAS summary statistics, and differential expression. (a) FDR values (Fisher’s exact test) for enrichment of de novo loss-of-function variants from ASD and SCZ within modules, summarized to module sets. Bar height gives geometric mean of FDR, and whiskers the range of significant FDR values for modules within the module set. Modules with bold labels on x-axis show enrichment from GWAS summary statistics (see b, Methods). Module sets are ordered by Jaccard similarity between their index modules. Green region: These modules enrich for neuronal markers. Blue region: These modules enrich for fetal neuron, mitotic progenitor, or outer radial glia markers. (b) FDR values (MAGMA; Methods) for GWAS summary statistics within modules. Method of ordering identical to (a). (c) Standard boxplot (box: quartiles, whiskers: 1.5xIQR) of module eigengene expression for BW-M1 and BW-M4 in ASD cases and control brains across three regions and associated p-values from a prior ASD sequencing study. P-values: T-test (unsided) (d-f). Volcano plots for individual genes in modules BW-M1 (NPC), BW-M6 (astrocyte) and BW-M4 (neuron) in a prior ASD sequencing study; x-axis: log fold-change, y-axis: p-value (linear mixed model), sign-test p-values are inset demonstrating an overabundance of up-regulated genes in BW-M1, BW-M6, and down-regulated genes in BW-M4 (Methods) (g) Module preservation statistics for ASD and controls calculated separately for cortical and cerebellar modules shows highly consistent patterns, except for CTX-M3 and CEREB-M1, which are differentially preserved.