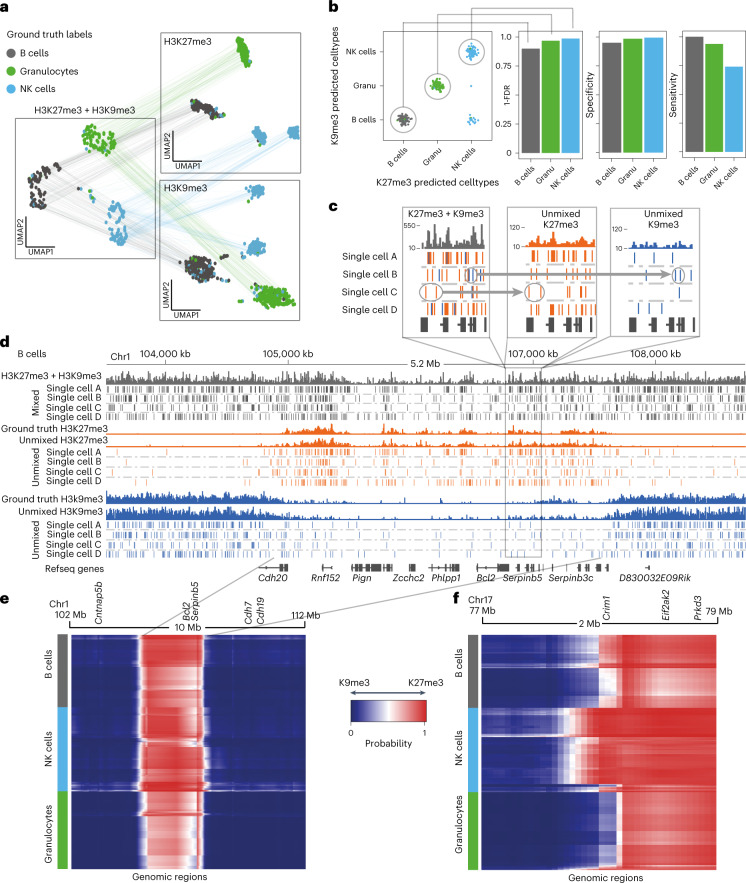
Fig. 2. scChIX-seq accurately deconvolves multiplexed histone modifications in single cells.
a, UMAP representation of the H3K27me3 (n = 367) and H3K9me3 (n = 376) histone modification space derived from the two single-incubated datasets (right two panels), and the H3K27me3+H3K9me3 space (left panel, n = 290) derived from the double-incubated data. Cells are colored by their ground truth cell-type labels. The cells in the H3K27me3- and H3K9me3-only space have unmixed double-incubated cells whose deconvolved signal has been projected onto their respective UMAPs. Lines connecting across datasets connect where each double-incubated cell is located in each of the three histone modification space. b, Matrix summarizing the cluster pair that scChIX-seq selected for each double-incubated cell. Cells along the diagonal are predicted to be B cells, granulocytes and NK cells, respectively. Cells in the off-diagonal are false negatives. Barplots summarizing FDR, sensitivity and specificity of assigning each cell type (right). c, Zoom-in coverage plot and single-cell cut fragments in B cells of mixed (H3K27me3+H3K9me3, gray bars), unmixed (H3K27me3 and H3K9me3, orange and blue bars). Positions of cut fragments are shown for four single cells (single cells A, B, C and D) for H3K27me3+H3K9me3 signal (gray ticks) as well as their unmixed outputs (orange and blue ticks). Circled reads and arrow highlight examples of cut fragments being assigned to either H3K27me3 (orange) or H3K9me3 (blue). d, Zoom-out of the Serpinb5 locus. Cut fragments from H3K27me3+H3K9me3 are colored based on whether they have been assigned to H3K27me3 (orange) or H3K9me3 (blue). Ground truth coverage are single-incubated sortChIC data targeting H3K27me3 (orange) and H3K9me3 (blue). e, Heatmap of probabilities p of assigning reads to H3K27me3 (p = 1, red) or H3K9me3 (p = 0, blue) around the Bcl2 locus. Rows are single cells (ordered by predicted cell type), columns are genomic regions (50 kb bins). Transitions between H3K9me3- and H3K27me3-marked chromatin states are independent of cell type. f, Same as e but at the Crim1 locus, where transitions from H3K9me3 to H3K27me3 (blue to red) are cell-type specific.