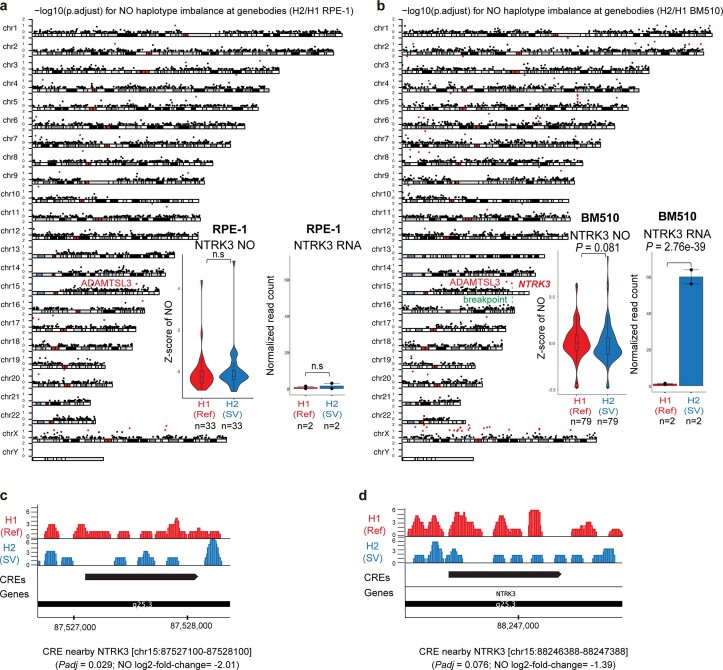
Extended Data Fig. 5. Haplotype-specific NO analysis in RPE-1 and BM510.
(a, b) Haplotype-specific NO analysis of NO at gene bodies genome-wide in RPE-1 (a) and BM510 (b). For each chromosomal karyogram, the y-axis indicates the significance of haplotype-specific NO for each gene (-log10 p.adjust). All the significant genes were indicated in red dots (FDR 10%; two-sided wilcoxon rank sum test followed by Benjamini Hochberg multiple correction; derived from n = 33 cells and n = 79 cells for RPE-1 and BM510, respectively; Boxplots were defined by minima = 25th percentile - 1.5X interquartile range (IQR), maxima = 75th percentile + 1.5X IQR, center = median, and bounds of box = 25th and 75th percentile.). NTRK3 (identified in BM510) is the only significant gene adjacent to an SV breakpoint. Haplotype-resolved RNA expression at the NTRK3 locus is depicted using bar graphs in the right panel (two-sided likelihood ratio test followed by Benjamini Hochberg multiple correction; n = 2 biological replicates; Data are presented as mean values +/− SEM). (c-d) Haplotype-specific NO analysis at CREs. Browser track depicts the haplotype-resolved NO of the not rearranged (Ref) homolog in red, and the SV homolog in blue. scNOVA identified two CREs with significant haplotype-specific NO, including an intergenic CRE spanning chr15:87527100-87528100 (p.adjust = 0.029, log2-fold change = −2.01) (c) and an intronic CRE at chr15:88246388-88247388 (p.adjust = 0.076, log2-fold change = −1.39) (d).