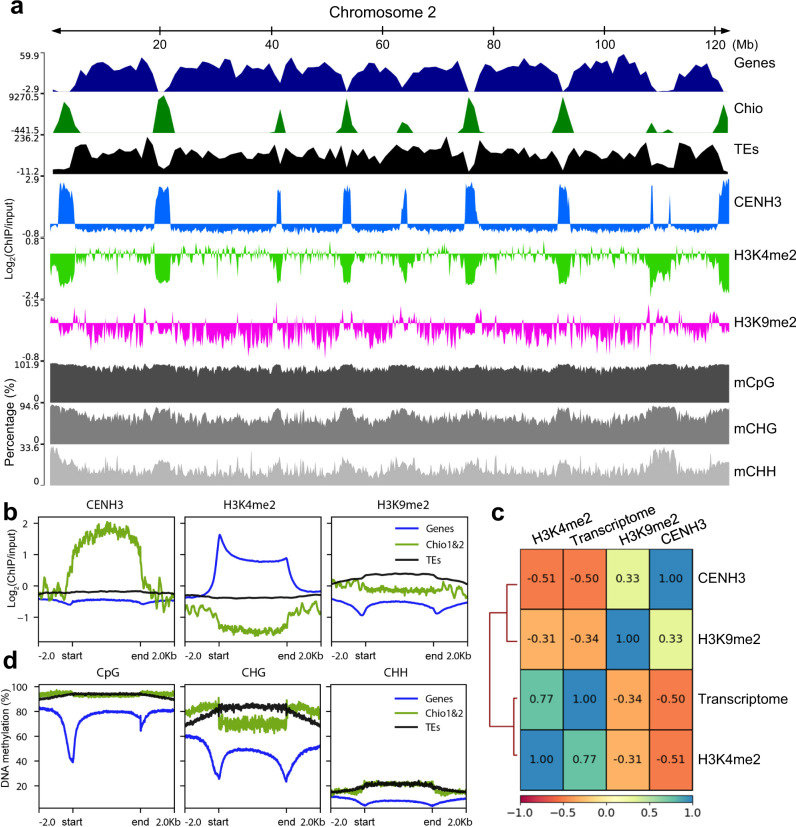
Fig. 5. The genome of C. japonica is organized in large-scale eu- and heterochromatic regions.
a Distribution of annotated genes, high-copy centromeric Chio satellite repeats, and transposable elements (TEs), as well as the enrichment of CENH3, H3K9me2, and H3K4me2 ChIPseq. The ChIPseq signal tracks are represented as the average log2 ratio of ChIP/input in genome-wide 10 kb windows. Chromosome 2 is taken as a representative chromosome. DNA methylation level at CpG, CHG, and CHH is shown in percentage. b Genome-wide enrichment of CENH3, H3K4me2, and H3K9me2 at different types of sequences, including genes (blue), centromeric Chio satellite arrays (green), and TEs (black). c The heatmap shows the correlation scores among different ChIPseq samples (CENH3, H3K9me2 and H3K4me2) and transcriptome. d The methylation level of CpG, CHG, and CHH at genes (blue), Chio arrays (green), and TEs (black).