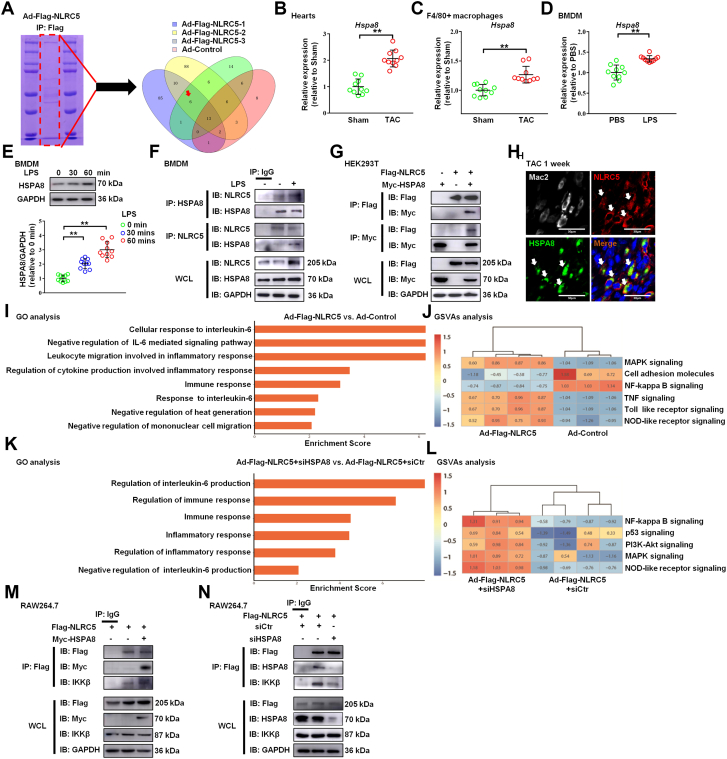
Figure 5.
NLRC5 Directly Interacted With HSPA8
(A) The most potential functional binding targets of NLRC5 were evaluated by mass spectrometry of BMDMs with adenovirus (Ad) overexpressing NLRC5. Six potential target proteins were interacting with NLRC5 were screened through 3 batches of overexpressed NLRC5 compared with the result of a control group. (B) Relative mRNA expression of Hspa8 in hearts after TAC. (C) Relative mRNA expression of Hspa8 in F4/80+ macrophages isolated from hearts. (D) Relative mRNA expression of Hspa8 in BMDMs with or without LPS treatment. (E) Western blotting and quantitative analysis of HSPA8 and GAPDH in BMDMs from mice with or without LPS treatment. (F) Coimmunoprecipitation of NLRC5 and HSPA8 in BMDMs with and without LPS treatment. The lysates immunoprecipitated with anti-IgG serve as a negative control condition. (G) HEK293T cells were cotransfected with Flag-tagged NLRC5 and Myc-tagged HSPA8 plasmid. (H) Immunofluorescence staining of NLRC5 (red), Mac2 (grey), HSPA8 (green), and nuclei (blue) in hearts. Scale bars, 30 μm. (I) Gene ontology (GO) enrichment analysis and gene set variation analysis (GSVAs) of the expression of several genes with roles in different biological responses of Ad-Flag-NLRC5 compared with the Ad-negative control (Ad-NC) condition from RNA sequencing. (J) GSVAs of the expression of several genes were performed with role in different signaling pathways from RNA sequencing. (K and L) GO enrichment analysis and GSVAs analysis of the expression of several genes with role in different biological response and signaling pathways of Ad-Flag-NLRC5 plus siHSPA8 treatment compared with Ad-Flag-NLRC5 from RNA sequencing. (M) BMDMs were cotransfected with Flag-tagged NLRC5 and Myc-tagged HSPA8 plasmid. The lysates immunoprecipitated with anti-IgG serve as a negative control condition. (N) Coimmunoprecipitation of NLRC5 and IKKβ in BMDMs with or without siHSPA8 treatment. Data are expressed as mean ± SD. ∗P < 0.05, ∗∗P < 0.01. Data in B to D were analyzed using Student’s unpaired t-test. Data in panel E were analyzed using 1-way analysis of variance followed by Tukey’s post hoc analysis. IB = immunoblotting; IP = immunoprecipitation; WCL = whole cell lysis; other abbreviations as in Figures 1 and 2.