Figure 4.
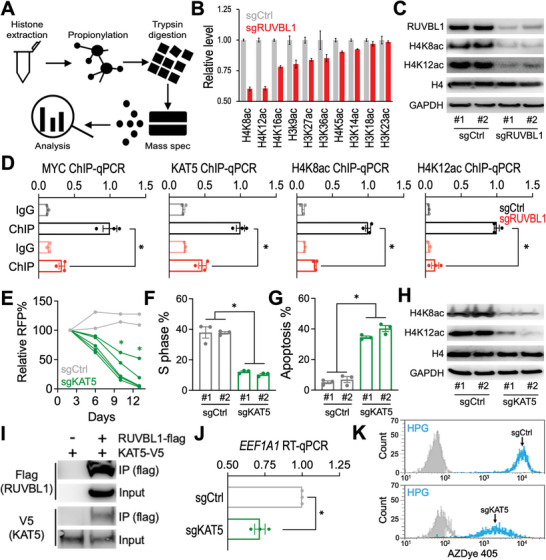
RUVBL1 modulates histone H4 acetylation via KAT5. A) Schematic outline of histone modification mass spectrometry and B) levels of histone H3 and H4 acetylation detected in sgCtrl and sgRUVBL1 transduced A673‐Cas9 cells (n = 3 mass spec measurements per sample). C) Western blot of RUVBL1, H4K8ac, H4K12ac, histone H4, and GAPDH in A673‐Cas9 cells transduced with sgCtrl versus sgRUVBL1 (two independent sgRNA sequences per group). D) ChIP‐qPCR of MYC, KAT5, H4K8ac, and H4K12ac at EEF1A1 locus in A673‐Cas9 cells transduced with sgCtrl and sgRUVBL1 (n = 3 for each group). E) Growth competition assay of sgCtrl (gray lines; n = 2 independent sgRNA sequences) and sgKAT5 (green lines; n = 5 independent sgRNA sequences) in A673‐Cas9 cells. F) Cell cycle monitored by EdU incorporation, and G) cellular apoptosis detected by active caspase 3+/DAPI− in A673‐Cas9 cells transduced with sgCtrl and sgKAT5 (n = 3 for each group). H) Western blot of RUVBL1, H4K8ac, H4K12ac, histone H4, and GAPDH in A673‐Cas9 cells transduced with sgCtrl versus sgKAT5 (two independent sgRNA sequences per group). I) Co‐IP of RUVBL1 (flag‐tagged) with KAT5 (V5‐tagged) in HEK293 cells. J) RT‐qPCR of EEF1A1 mRNA in A673‐Cas9 cells transduced with sgCtrl and sgKAT5 (n = 3 for each group). K) Flow cytometric profiles of HPG labeled (cyan) compared to the nonlabeled (gray) cells in A673‐Cas9 cultures transduced with sgCtrl versus sgKAT5. Data are represented as mean ± SEM. *P < 0.01 compared to sgCtrl by two‐sided Student's t‐test. Source data are available for this figure: SourceData F4 B.
