Figure 2.
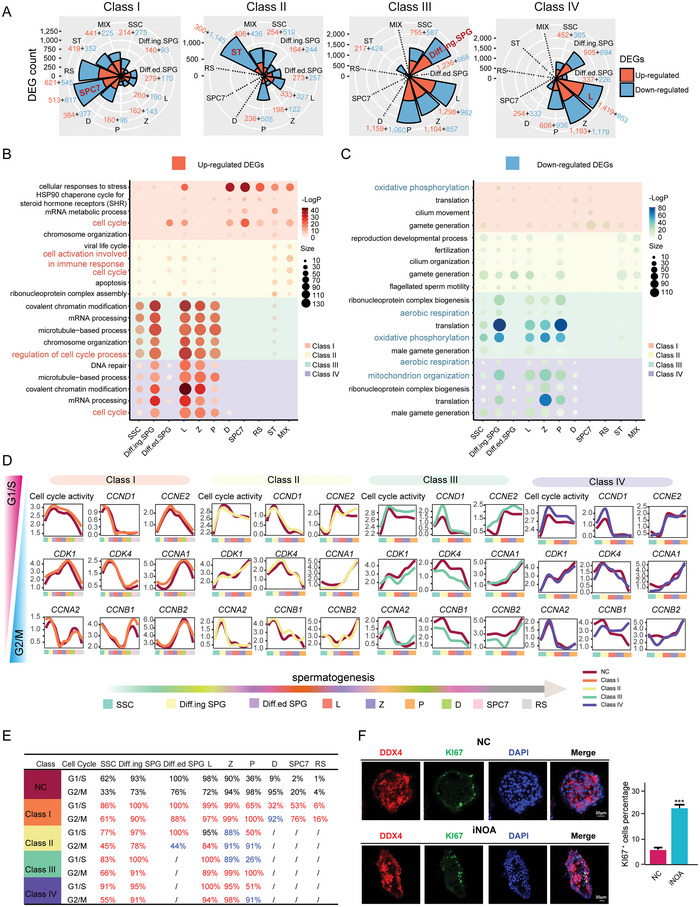
Transcriptional features of the four classes idiopathic nonobstructive azoospermia (iNOA). A) differentially expressed genes (DEGs) between normal controls (NC) and the four classes iNOA. B) Functions of the upregulated DEGs in the four classes iNOA. C) Functions of the downregulated DEGs in the four classes iNOA. D) Expression levels of G1/S and G2/M phase genes in the four classes iNOA. E) The ratio of cells with active cell cycle to total cells in each cluster according to the expression of G1/S and G2/M phase genes. F) Left panel, immunofluorescence of DDX4 (red, a marker for germ cell) costained with KI67 (green) in NC and iNOA. The scale bar represents 20 µm. Right panel, bar plot of the KI67 + cell percentage. A two‐tailed Student's t test was used for the significance test.
