FIGURE 3.
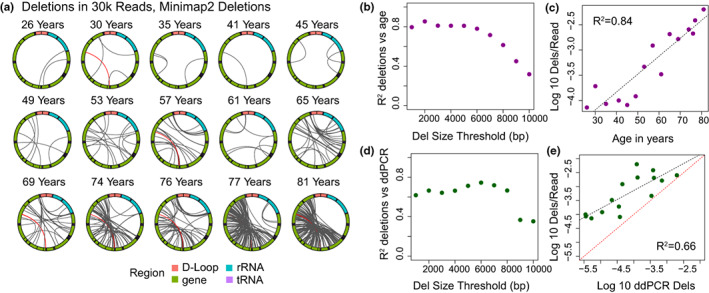
Large mtDNA deletions mapped using combined Minimap2 alignments. (a) Localization of deletions >2000 bp identified using the Minimap2 algorithm in a random subset of 30,000 reads per sample. The mitochondrial genome is depicted in a clockwise orientation with lines linking the start and end of each identified deletion. The human mitochondrial “common” deletion is highlighted in red where it was detected. (b) R 2 correlation coefficient for log‐transformed number of identified deletions per mitochondrial read versus age plotted versus the minimum deletion size cutoff used in calculating correlation. (c) Log‐transformed frequency of deletions >2 kbp per mitochondrial read versus age. (d) R 2 correlation coefficient for log‐transformed number of identified deletions per mitochondrial read versus log‐transformed deletion frequency by ddPCR plotted versus the minimum deletion size cutoff used in calculating correlation. (e) Log‐transformed frequency of deletions >2 kbp per mitochondrial read versus log‐transformed ddPCR deletion frequency. Red dashed line is the line of identity.
