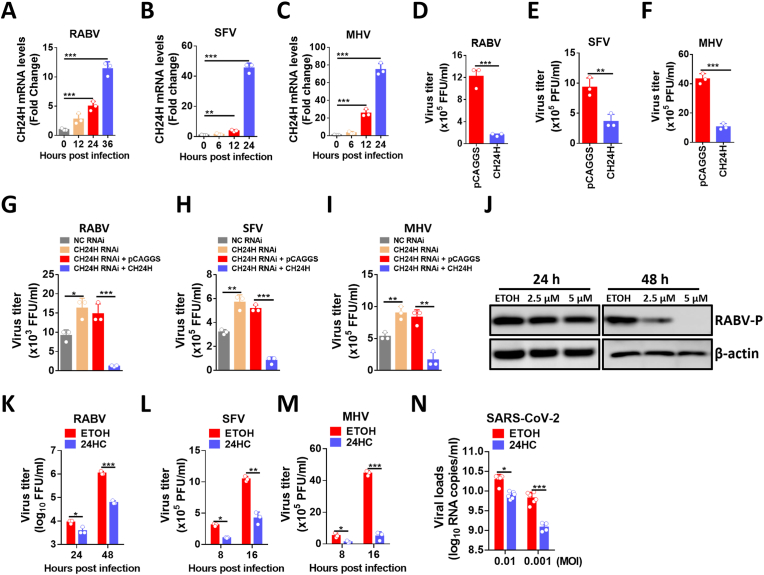
Fig. 7.
CH24H and 24HC restrict the replication of several viruses. A-C. N2a cells were infected with RABV (A) at an MOI of 0.01, SFV (B) at an MOI of 0.01, or MHV (C) at an MOI of 0.01, and cells were harvested at the indicated time points. CH24H mRNA levels were measured by qPCR (n = 3). D-F. N2a cells were transfected with pCAGGS vector or pCAGGS-CH24H for 24 h, and then cells were infected with RABV (D) at an MOI of 0.01 for 48 h, SFV (E) at an MOI of 0.01 for 16 h, or MHV (F) at an MOI of 0.01 for 16 h. The supernatants were collected for virus titration (n = 3). G-I. N2a cells were transfected with negative control siRNA (NC RNAi), CH24H-specific siRNA (CH24H RNAi), pCAGGS vector with CH24H RNAi, or CH24H with CH24H RNAi. After 24 h incubation, cells were infected with RABV (G) at an MOI of 0.01 for 24 h, SFV (H) at an MOI of 0.01 for 8 h, or MHV (I) at an MOI of 0.01 for 8 h. Cell supernatants were collected for virus titration (n = 3). J, K. N2a cells were pretreated with 24HC at the indicated concentrations (2.5 μM or 5 μM) for 12 h prior to RABV infection at MOI 0.01, the cells were harvested at 24 and 48 h post-infection. WB (J) and virus titration (K) were performed to assess viral replication levels (n = 3). L. N2a cells were pretreated with 5 μM 24HC for 12 h, and then infected with SFV at MOI 0.01 for 8 or 16 h. Cell supernatants were collected for virus titration (n = 3). M. N2a cells were pretreated with 5 μM 24HC for 12 h, and then infected with MHV at MOI 0.01 for 8 or 16 h. Cell supernatants were collected for virus titration (n = 3). N. MDCK cells were pretreated with 5 μM 24HC for 6 h, and then infected with SARS-CoV-2 at the indicated MOIs for 24 h. Cell supernatants were collected and qPCR was performed to detect SARS-CoV-2 viral load (n = 6). Statistical analysis of grouped comparisons was performed by Student's t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Bar graph represents mean ± SD. Western blot data are representative of at least two independent experiments.