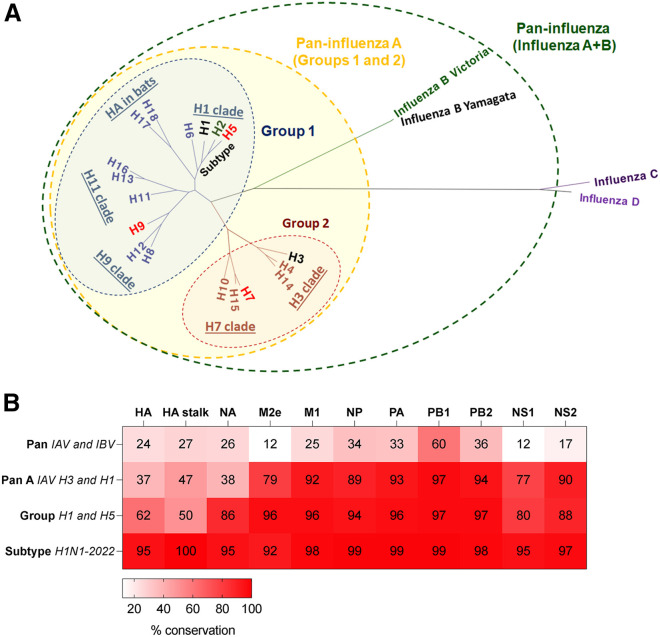
Figure 2.
Sequence conservation among different influenza virus clades
(A) Definition of influenza breadth based on HA phylogeny. The tree was built with IQ-TREE2 (default parameters with automatic model selection) using HA (HE) protein sequences obtained from the Influenza Virus Database. Human influenza viruses as indicated (black: subtype endemic to humans, green: (H2, B/Yam) previously in humans, red: spill over infections from zoonotic sources).
(B) Amino acid sequence conservation based on sequence alignment (CLUSTALWjp) of proteins (from https://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi?go=genomeset) of H1N1 2009 pandemic (A/California/04/2009) versus prototypic virus strains: for Pan (B/Vic, B/Brisbane/60/2008), for Pan A (H3N2, A/Alaska/03/2021), for group 1 (H5N1, A/Indonesia/5/2005), for subtype (H1N1, A/Alabama/01/2020).