Fig. 1.
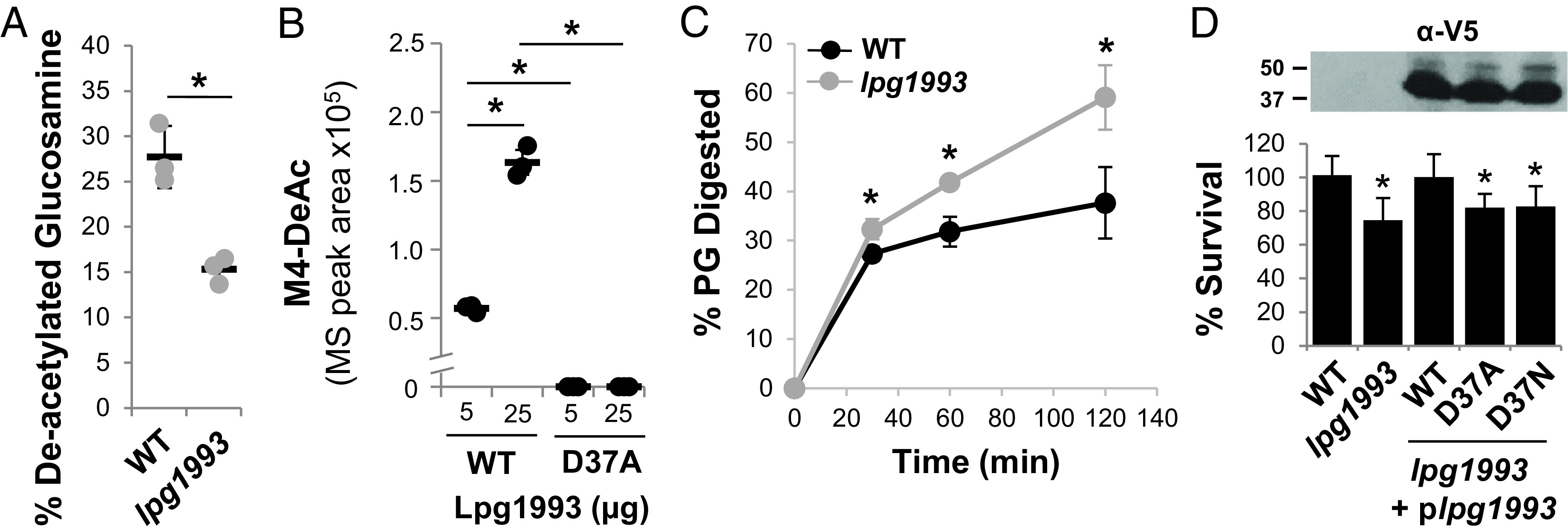
Lpg1993 catalyzes the deacetylation of N-acetylglucosamine protecting L. pneumophila peptidoglycan from lysozyme digestion. (A) lpg1993 encodes an N-linked N-acetylglucosamine deacetylase. Quantitative structural analysis of peptidoglycan muropeptides of wild-type (WT) and Δlpg1993 bacteria. (B) Lpg1993 catalyzes the deacetylation of peptidoglycan in vitro. Varying amounts of wild-type (WT) or catalytically inactive (D37A) Lpg1993 protein were incubated with purified peptidoglycan, and the amount of NAG deacetylation based on the abundance of deacetylated GlcN-MurNAc-tetrapeptide (M4-DeAc) was measured by UPLC-MS. (C) Peptidoglycan of Δlpg1993 bacteria is more sensitive to lysozyme digestion. Peptidoglycan isolated from WT and Δlpg1993 bacteria was treated with lysozyme, and the percentage of hydrolyzed peptidoglycan was measured by UPLC-MS. (A–C) Data are the mean ± SD of 3 biological replicates. (D) Lpg1993 protects L. pneumophila against lysosome-mediated killing. (Top) Western analysis of whole-cell lysates of WT and Δlpg1993 bacteria harboring empty vector or Δlpg1993 bacteria expressing V5-6×HIS dual epitope–tagged fusion proteins of wild-type Lpg1993 (WT) or variants lacking the conserved catalytic residue aspartate residue (D37A, D37N) grown to post-exponential phase. Data are representative of 3 biological replicates. (Bottom) Lysozyme sensitivity assay. Bacteria were treated with lysozyme, and percent survival was measured based on recovered colony-forming units (cfus) on solid medium comparing lysozyme treatment to no lysozyme control. Data are the mean ± SD of 6 biological replicates, each consisting of 3 technical replicates. (A–D). An asterisk indicates a Student’s t test P < 0.05 relative to the WT strain, unless otherwise indicated.
