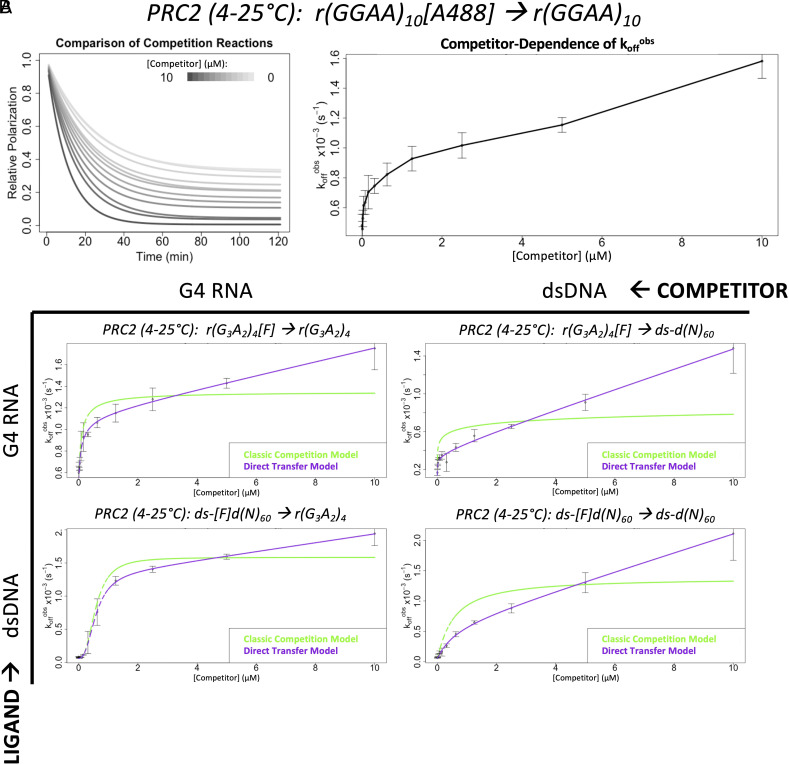
Fig. 2.
PRC2 exhibits direct transfer kinetics for G4 RNA and dsDNA. FPCD experiments (Fig. 1) were performed with the Wang et al. and Long et al. ligand/competitor over a range of competitor concentrations (panel A) and to measure direct transfer kinetics for every ligand–competitor combination of a G4 RNA and 60-bp dsDNA (panel B). Data are from representative experiments (of n ≥ 3), where error bars indicate mean ± SD for four technical replicates. Rate constant values from regression can be found in Table 1, additional nomenclature definitions are in SI Appendix, Table S1, and polynucleotide species definitions are in SI Appendix, Table S2. (A) Exponential regression fit lines from each condition (left plot), alongside the observed initial dissociation rates (koffobs, see SI Appendix, Eq. S3.2) as a function of competitor concentration (right plot). Solid line in right plot is a visual aid connecting data means. Raw data are shown in SI Appendix, Fig. S1. (B) Experiments were performed in BB10 buffer. Isotherm, carrier nucleic acid, and fluorophore controls for the RNA-RNA competition experiment (Top Left) can be found in SI Appendix, Fig. S2. Analogous studies with a 50-bp dsDNA can be found in SI Appendix, Fig. S3.