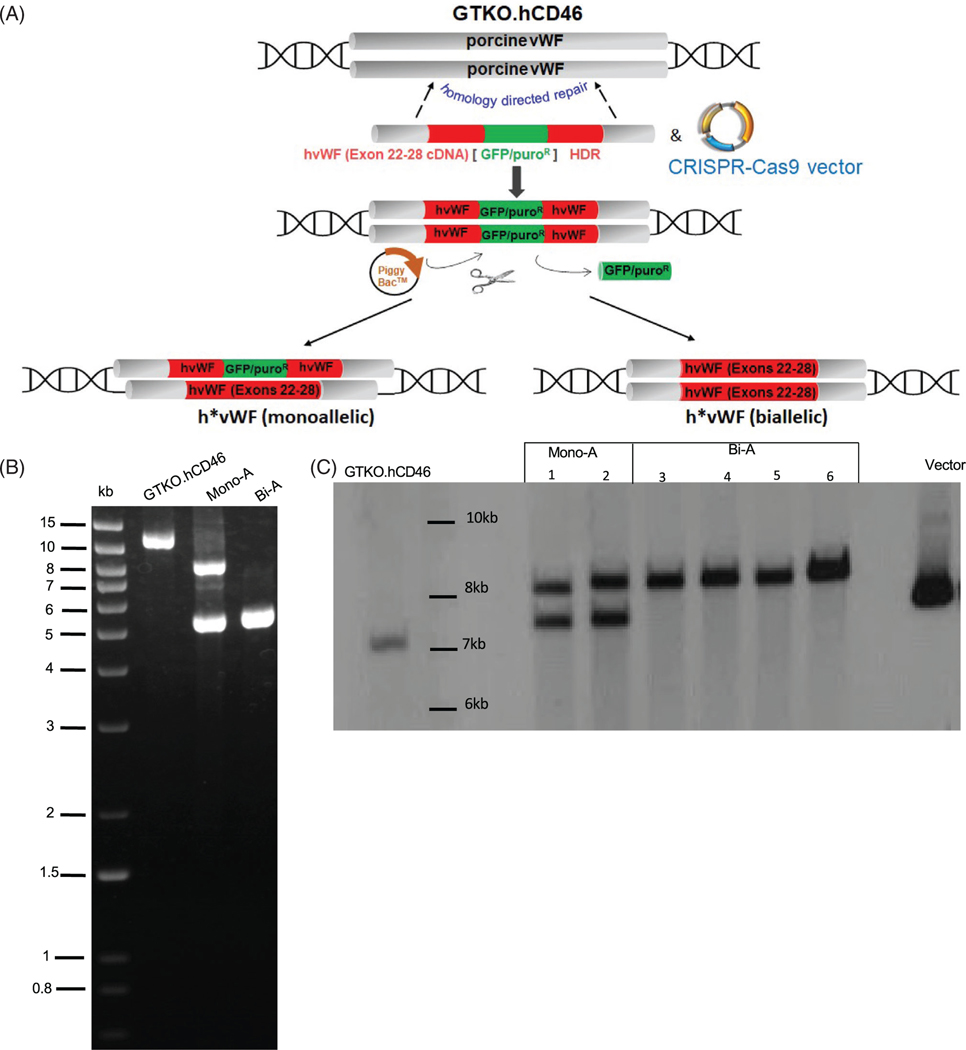
FIGURE 1.
Generation of humanized pig von Willebrand factor (vWF) by homology directed repair. (A) GTKO.CD46 porcine fetal fibroblasts were transfected with a targeting vector containing cDNA of human vWF (hvWF) exons 22–28 and a transposase-excisable GFP/PuroR selectable marker and plasmids expressing target-specific CRISPR-Cas9. Single cell clones of targeted fibroblasts were subjected to somatic cell nuclear transfer (SCNT), followed by embryo transfer to recipient pigs from which fetuses were collected on gestation Day 32. Fibroblasts from these fetuses were transfected with transposase-expressing plasmids to excise the selectable marker and subjected to a second round of SCNT to generate pigs. (B) Long-range polymerase chain reaction (PCR) on genomic DNA of GTKO.hCD46 and vWF-edited pigs. Primers are located just outside of the 5′ and 3′ homology arm sequences. The wild type (WT) allele produces a band of 11.7 kb, while the monoallelic (Mono-A) modification produces two bands, one of 8.2 kb, which contains the “humanization of pvWF (h*pvWF) insert plus the unexcised GFP/PuroR selectable marker, and another of 5.3 kb produced by the allele in which the marker was excised. The bi-allelic modification (Bi-A) produces a single band of 5.3 kb. This was sequenced to confirm precise integration of hvWF exons 22–28 in pvWF and precise excision of the selectable marker. (C) Southern blot was performed by digesting genomic DNA with HindIII/Psil restriction enzymes and using a DIG-labeled hvWF probe (exon 24–28, with partial sequence homology to pig vWF). pvWF generates a single band of 7 kb; Mono-A generates two bands of 7.5 kb and 8.4 kb, representing alleles in which the GFP/PuroR marker was unexcised or excised, respectively; and Bi-A generates a single band of 8.4 kb. Targeting vector serves as a positive control