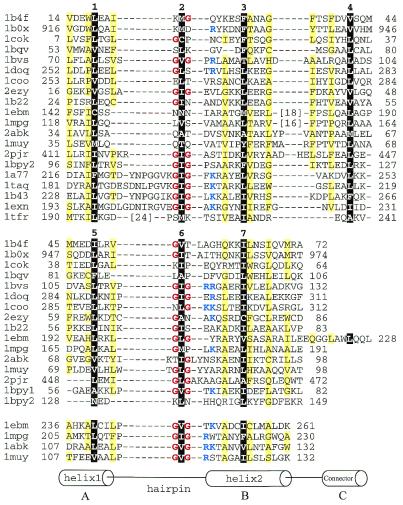
Figure 1.
Structure-based sequence alignment of HhH proteins. For each sequence, PDB entry name and starting and ending residue numbers are given. Protein name, chain name (if any) and gene identifier (gi) number for each entry are: 1b4f, human EphB2 receptor, chain A, 4558093; 1b0x, mouse EphA4 receptor tyrosine kinase, chain A, 4929864; 1cok, human p73 C-terminal domain, chain A, 5822025; 1bqv, mouse Ets-a transcription factor pointed domain, 3891925; 1bvs, Mycobacterium leprae DNA-helicase RuvA middle domain, chain A, 3660156; 1doq, T.thermophilus RNA-polymerase α-subunit C-terminal domain, chain A, 6730428; 1coo, E.coli RNA-polymerase α-subunit C-terminal domain, 1421046; 2ezy, human BAF, chain A, 4389121; 1b22, human DNA repair protein Rad51 N-terminal domain, chain A, 6730074; 1ebm, human 8-oxoguanine glycosylase central domain, chain A, 2078294; 1mpg, E.coli 3-methyladenine DNA glycosylase II 2 C-terminal domains, chain A, 2914353; 2abk, E.coli endonuclease III, 1311214; 1muy, E.coli MutY catalytic domain, chain A, 5822134; 2pjr, Bacillus stearothermophilus DNA-helicase PcrA insertion domain, chain A, 4930184; 1bpy1 and 1bpy2, human DNA-polymerase beta N-terminal (8 kDa) domain and ‘fingers’ domain, respectively, chain A, 2392200; 1a77, Methanococcus jannaschii Flap endonuclease-1, 5821778; 1taq, Thermus aquaticus DNA-polymerase Taq 5′ to 3′ exonuclease domain, 1942938; 1b43, Pyrococcus furiosus Fen-1 nuclease, chain A, 6980604; 1exn, bacteriophage T5 5′-exonuclease, chain A, 2392326; 1tfr, bacteriophage T4 RNase H, 1943457. The first HhH motifs are aligned in the top panel. For proteins with more than one HhH motif, the second and the third motifs are aligned in the middle and the bottom panels, respectively. All three panels are also aligned with each other. Positions of hydrophobic core residues are highlighted in black and are labeled with numbers corresponding to those in Figure 2c. Residues in the third HhH motif are not numbered. Glycines in the signature sequence GhG (h is a hydrophobic residue) of the hairpin regions are in red and positively charged residues following the signature are in blue. Positions with mostly uncharged residues are shaded in yellow. Numbers in brackets indicate the number of omitted residues in the sequence.