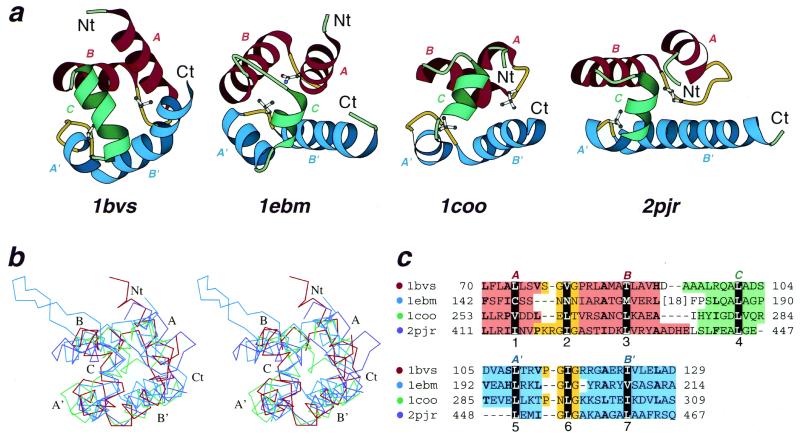
Figure 2.
Structural comparisons of divergent (HhH)2 domains. (a) Structural diagrams of DNA-helicase RuvA middle domain (1bvs, chain A, residues 63–134), 8-oxoguanine glycosylase central domain (1ebm, chain A, 135–221), C-terminal domain of RNA-polymerase α-subunit (1coo, 253–296) and DNA-helicase PcrA insertion domain (2pjr, chain A, 405–478), showing the HhH motifs from each protein. For each protein, N- and C-termini are labeled with Nt and Ct, respectively. The helices in the first HhH motif are labeled with A and B, and are in red. The corresponding helices in the second HhH motif are labeled with A′ and B′ and are in blue. The hairpin regions of both motifs are in yellow. Side-chains of central hydrophobic residues in hairpins are shown using ball-and-stick representation. The helices connecting the two HhH motifs are labeled with C and are in green. The ribbon diagrams were rendered by Bobscript (28), a modified version of Molscript (29). (b) Stereo diagram of superimposed Cα traces of DNA-helicase RuvA subunit (red), 8-oxoguanine glycosylase (blue), C-terminal domain of RNA-polymerase α-subunit (green) and DNA-helicase PcrA (purple). Superpositions were made using InsightII package (Molecular Simulations Inc.). Labels match those described in (a). (c) Structure-based sequence alignment of HhH motif regions of the four illustrated protein domains. For each sequence the PDB entry name and starting and ending residue numbers are given. The dot color scheme (in front of each PDB entry) matches those in (b). Color shading and helix labels correspond to those in (a). The two HhH motifs (upper and lower panels) are aligned with each other. Sites of conserved core hydrophobic residues are highlighted in black, and are labeled 1, 2 and 3 in the first HhH motif, 5, 6 and 7 in the second motif, and 4 in the connector helix C (also see Fig. 1). Additional conserved hydrophobic residues are shown in bold.