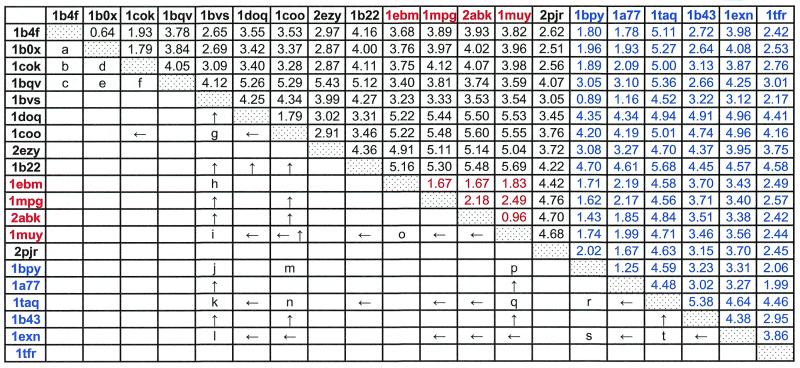
Table 1. Structural comparison among HhH motif-containing proteins and their sequence relations.
PDB entries of 20 representative HhH motif-containing proteins are listed in the top row and left column. The HhH domain proteins, containing two HhH motifs with a connector helix in between, are in black; those containing one additional HhH motif are in red; those that have only one HhH domain are in blue. Numbers in the upper right half of the table are r.m.s.d. values between each pair of proteins. Each pair of proteins was superimposed by Cα traces with InsightII package (Molecular Simulations Inc.). Numbers in black were obtained by superimposing the two HhH motifs and the connector helix; those in red by superimposing the same region plus the additional HhH motif; those in blue by superimposing only one HhH motif. See Materials and Methods for the definition of superimposed regions. The lower left half of the table shows PSI-BLAST search results. A letter indicates a single starting query sequence that can detect the proteins in the row and in the column in PSI-BLAST iterations with 0.01 as an E-value threshold before convergence (see Materials and Methods). The gi numbers:residue ranges of them are: a, 4929864:23_98; b, 4929864:23_98; c, 3386625:220_356; d, 4929864:23_98; e, 3386625:220_356; f, 3386625:220_356; g, 3560537:1_124; h, 3182983:477_593; i, 3182983:415_589; j, 2127858:1113_1192; k, 2127858:1113_1192; l, 586902:220_348; m, 42144:404_494; n, 6458154:80_175; o, 2982908:48_146; p, 4981594:11_181; q, 1651660:446_601; r, 549012:123_252; s, 4980590:436_598; t, 2983968:158_289. An arrow indicates that a sequence of one protein can find the other protein sequence through multiple rounds of PSI-BLAST iterations using all significantly different detected homologs as queries (see Materials and Methods) in subsequence iterations. An ‘up’ arrow shows that the protein in the row was used as a starting query to detect the protein in the column, and a ‘left’ arrow indicates the opposite situation.