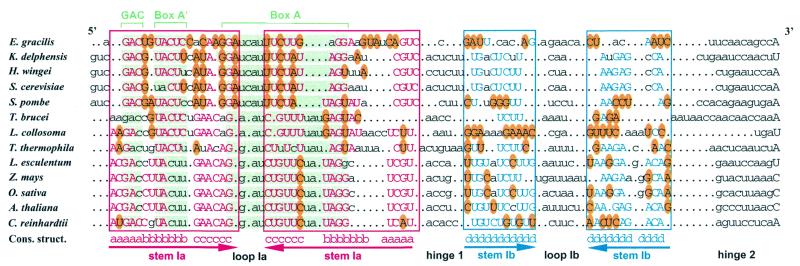
Figure 1.
Compensatory base mutations conserved the possibility of forming a two stem–loop structure in yeast, ciliate, unicellular algal and plant U3 snoRNAs. The sequences of the 5′-terminal domain from Euglena gracilis (Eg27297), Kluyveromyces delphensis (Kdz78433), Hansenula wingei (Hwu3sno), Saccharomyces cerevisiae (Scsnr17a), Schizosaccharomyces pombe (Spsnru3), Trypanosoma brucei (Tburb), Leptomonas collosoma (Lctgv3snr), Tetrahymena thermophila (Ttsnru31), Lycopersicon esculentum (Leu3snr), Zea mays (Zmu3snrng), Oryza sativa (Osu3snrn), Arabidopsis thaliana (Atu3csnr) and Chlamydomonas reinhardtii (Crj001179) were first subjected to the program developed by Lück et al. (31). Then alignment was manually refined for a better alignment of the phylogenetically conserved boxes GAC, A′ and A (highlighted in green) and for a better representation of co-variations. The nucleotide sequences involved in the stem of stem–loop Ia are boxed in red. Stems are marked below by inverted arrows. The base paired residues are in capital letters. Red capital letters correspond to residues fitting the consensus sequence established from the alignment. Nucleotide variations from this consensus sequence that were found to preserve base pair interactions are circled in yellow. Dots indicate missing nucleotides as referred to the alignment. The same representation is used for stem–loop Ib, except that blue is used instead of red. The sequences can be accessed via the EMBL-ID at http://srs.ebi.ac.uk:5000/site