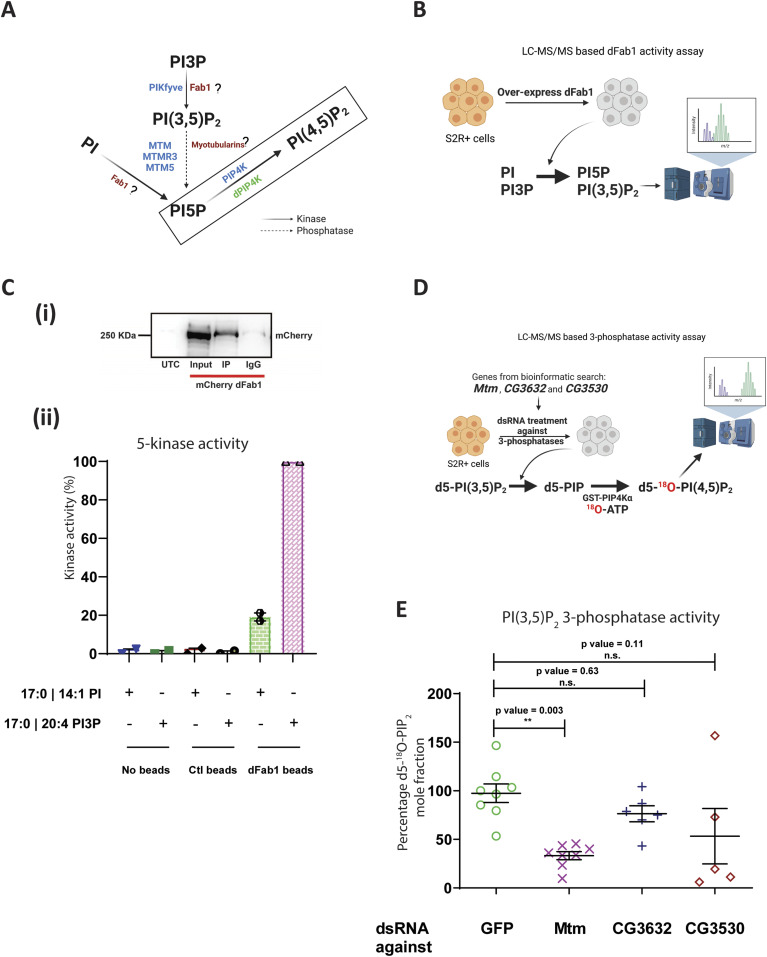
Figure 1. Screening for a biochemical route to modulate PI5P levels in Drosophila as altering local PI(4,5)P2 could not change the cell size of dPIP4K29.
(A) Schematic illustrating the putative enzymatic routes by which PI5P can be synthesised in Drosophila. The activities of enzymes labelled in blue are known in mammalian cells, the activity of enzymes labelled in red followed by “?” are still unknown in Drosophila, the activity of enzymes labelled in green are known in Drosophila. The activity of PI5P to PI(4,5)P2 is boxed and is linked to cell size regulation in Drosophila (Mathre et al, 2019). PI, phosphatidylinositol; PI3P, phosphatidylinositol 3-phosphate; PI(3,5)P2, phosphatidylinositol 3,5 bisphosphate; PI5P, phosphatidylinositol 5-phosphate; PI(4,5)P2, phosphatidylinositol 4,5 bisphosphate. Kinase and phosphatase activities are denoted by black solid arrows and black dashed arrows, respectively. (B) Schematic illustrating the LC-MS/MS-based in vitro 5-kinase activity assay using S2R+ cells overexpressing dFab1 enzyme to convert synthetic PI or PI3P to PI5P and PI(3,5)P2, respectively. (C) (i) Immunoprecipitated protein levels were analysed by Western blotting with an anti-mCherry antibody. Control (IgG) was prepared without anti-mCherry. Input lane shows the correct size of dFab1 protein ∼230 kD. UTC, Untransfected control. (ii) In vitro kinase assay on synthetic PI and PI3P. Graph representing the kinase activity (%) as the normalised response ratio of “PI 5-kinase activity on PI” to “PI3P 5-kinase activity on PI3P” upon enzymatic activity of immunoprecipitated mCherry::dFab1 on the respective substrates. Response ratio of PI 5-kinase activity on PI is obtained from area under the curve of 17:0 14:1 PI5P (product)/17:0 14:1 PI (substrate), Response ratio of PI3P 5-kinase activity on PI3P is obtained from area under the curve of 17:0 20:4 PI(3,5)P2 (product)/17:0 20:4 PI3P (substrate) and is represented as mean ± S.E.M. on addition of either negative control (no beads), control (mCherry beads) or dFab1 (mCherry::dFab1 beads). Number of immunoprecipitated samples = 2. (D) Schematic illustrating the LC-MS/MS-based in vitro PI(3,5)P2 3-phosphatase activity assay using dsRNA-treated S2R+ cells as the enzyme source to convert synthetic PI(3,5)P2 [d5-PI(3,5)P2 to d5-18O- PIP2] using a two-step reaction scheme. (E) In vitro phosphatase assay on synthetic PI(3,5)P2. Graph representing the 3-phosphatase activity (%) as the percent formation of d5-18O-PIP2 formed from starting d5-PI(3,5)P2 as mean ± S.E.M. on addition of either control (GFP ds RNA) or Mtm, CG3632, CG3530 ds RNA-treated S2R+ cell lysates. One-way ANOVA with a post hoc Tukey’s test shows P-value = 0.003 between GFP and Mtm ds RNA-treated lysates, shows P-value = 0.63 between GFP and CG3632 ds RNA-treated lysate and shows P-value = 0.11 between GFP and CG3530 ds RNA-treated lysates.