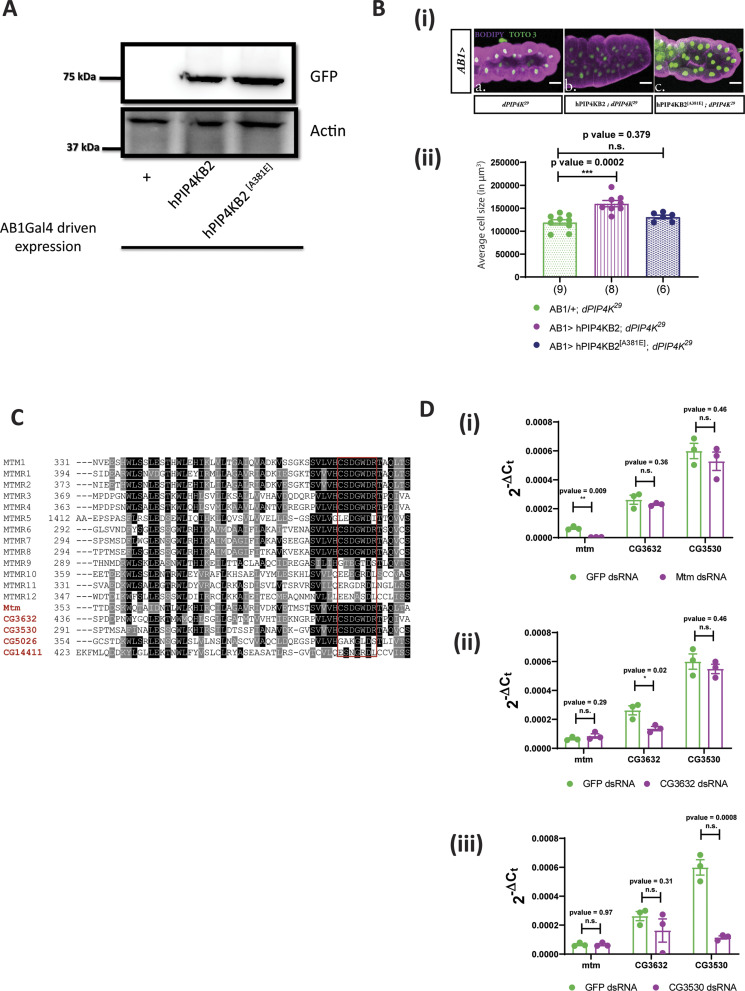
Figure S1. Fab1 and Mtm are potential PI5P-modulating enzymes in Drosophila.
(A) Protein levels between AB1/+; dPIP4K29 (Ctl), AB1 >hPIP4KB2::GFP; dPIP4K29 and AB1 >hPIP4KB2A381E::GFP; dPIP4K29 from salivary glands of third instar wandering larvae seen on a Western blot probed by GFP antibody. Both hPIP4KB2::GFP and hPIP4KB2A381E::GFP migrates at ∼75 kD. Actin was used as the loading control. (B) (i) Representative confocal images of salivary glands from the genotypes a. AB1/+; dPIP4K29, b. AB1>hPIP4KB2; dPIP4K29, c. AB1>hPIP4KB2[A381E]; dPIP4K29. Cell body is marked magenta by BODIPY conjugated lipid dye, and the nucleus is marked by TOTO-3 shown in green. Scale bar indicated at 100 μm. (ii) Graph representing average cell size measurement (in μm3) as mean ± S.E.M. of salivary glands from wandering third instar larvae of AB1/+; dPIP4K29 (n = 9), AB1>hPIP4KB2; dPIP4K29 (n = 8), AB1>hPIP4KB2[A381E]; dPIP4K29 (n = 6). Sample size is represented on individual bars. One-way ANOVA with post hoc Tukey’s test showed P-value = 0.0002 between AB1/+; dPIP4K29, and AB1>hPIP4KB2; dPIP4K29 and P-value = 0.379 between AB1/+; dPIP4K29, and AB1>hPIP4KB2[A381E]; dPIP4K29. (C) Multiple sequence alignment of the myotubularin phosphatase family proteins in Drosophila with human myotubularins share a common signature phosphatase catalytic motif, the C(X5)R motif (highlighted in red box) except CG5026, CG14411. The alignment was generated using clustalO and representation is using Jalview. Conservation is shown in range of white to black, black being most conserved. (D) (i) qPCR measurements for mRNA levels of Mtm, CG3632, and CG3530 from either GFP ds RNA- (green) or Mtm ds RNA-treated samples (magenta). Unpaired t test showed P-value = 0.009 between GFP ds RNA and Mtm ds RNA for Mtm, whereas P-value = 0.36, P-value = 0.46 for genes CG3632 and CG3530, respectively. (ii) qPCR measurements for mRNA levels of Mtm, CG3632, and CG3530 from either GFP ds RNA- (green) or CG3632 ds RNA-treated samples (magenta). Unpaired t test showed P-value = 0.02 between GFP ds RNA and CG3632 ds RNA for CG3632, whereas P-value = 0.29, P-value = 0.46 for genes Mtm and CG3530, respectively. (iii) qPCR measurements for mRNA levels of Mtm, CG3632 and CG3530 from either GFP ds RNA- (green) or CG3530 ds RNA-treated samples (magenta). Unpaired t test showed P-value = 0.0008 between GFP ds RNA and CG3530 ds RNA for CG3530, whereas P-value = 0.97, P-value = 0.31 for genes Mtm and CG3632, respectively.