Abstract
Background:
The importance of phosphorylation in sperm during spermatogenesis has not been pursued extensively. Testis-specific serine kinase 3 (Tssk3) is a conserved gene, but TSSK3 kinase functions and phosphorylation substrates of TSSK3 are not known.
Objective:
The goals of our studies were to understand the mechanism of action of TSSK3.
Materials and Methods:
We analyzed the localization of TSSK3 in sperm, used CRISPR/Cas9 to generate Tssk3 knockout (KO) mice in which nearly all of the Tssk3 open reading frame was deleted (ensuring it is a null mutation), analyzed the fertility of Tssk3 KO mice by breeding mice for four months, and conducted phosphoproteomics analysis of male testicular germ cells.
Results:
TSSK3 is expressed in elongating sperm and localizes to the sperm tail. To define the essential roles of TSSK3 in vivo, heterozygous (HET) or homozygous knockout (KO) male mice were mated with wild-type females, and fertility was assessed over four months; Tssk3 KO males are sterile, whereas HET males produced normal litter sizes. Absence of TSSK3 results in disorganization of all stages of testicular seminiferous epithelium and significantly increased vacuolization of germ cells, leading to dramatically reduced sperm counts and abnormal sperm morphology; despite these histologic changes, Tssk3 null mice have normal testis size. To elucidate the mechanisms causing the KO phenotype, we conducted phosphoproteomics using purified germ cells from Tssk3 HET and KO testes. We found that proteins implicated in male infertility, such as GAPDHS, ACTL7A, ACTL9, and REEP6, showed significantly reduced phosphorylation in KO testes compared to HET testes, despite unaltered total protein levels.
Conclusions:
We demonstrated that TSSK3 is essential for male fertility and crucial for phosphorylation of multiple infertility-related proteins. These studies and the pathways in which TSSK3 functions have implications for human male infertility and non-hormonal contraception.
Keywords: Knockout mouse, proteomics, oligozoospermia, teratozoospermia, oligoteratozoospermia
INTRODUCTION
Oligoteratozoospermia, a cause of idiopathic infertility in men, is a failure of spermatogenesis leading to both a decrease in sperm counts (oligozoospermia) and the presence of abnormal sperm forms (teratozoospermia).1 Genetic factors are considered to be one of the causes of non-obstructive oligozoospermia.2 Multiple genes responsible for azoospermia have been reported to date,3 and the utilization of this information for the development of infertility diagnosis,4,5 or the development of novel contraceptives targeting crucial reproductive tract-specific proteins,6,7 holds promise. Spermatogenesis, which takes place in the seminiferous tubule in the testis, demands a well-organized process in which spermatogonia undergo mitosis, meiosis, and spermiogenesis.8 During spermiogenesis, round spermatids are dramatically restructured into a suitable morphology for proper fertilization. Spermiogenesis is further divided into the three main phases: Golgi, acrosome cap/elongation, and maturation phases.9 Phosphorylation is a post-translational modification of proteins that is required for many cellular processes, including cell cycle progression, motility, metabolism, cell growth, and differentiation.10 The occurrence of phosphorylation is critical for proper sperm function as it is for other somatic cells. Protein phosphorylation in mammalian sperm motility and metabolic systems has been documented.11–14 Several studies support the importance of phosphorylation by kinases in spermatogenesis, but little is known about the mechanisms.15–17
The testis-specific serine kinase (TSSK) protein family members were cloned as kinase-encoding genes with restricted expression to mouse testis. Each family member shares a similar, serine/threonine protein kinase catalytic domain. There are six members in the TSSK family: TSSK1, TSSK2, TSSK3, TSSK4, and TSSK6 are conserved in mouse and human, while TSSK5 is protein coding in the mouse and is a pseudogene in human.16 Some of the gene knockout (KO) mouse models have been created and reported to affect male fertility. Tssk1/Tssk2 double KO and Tssk6 single KO mice display infertility, while Tssk4 single KO mice display subfertility.18–20 However, detailed functions and substrates of these kinases remain unknown.
Investigation of potential substrates of kinases is important to understand the function and significance of the target kinases, and for future applications. Identification of the substrates of TSSKs has been approached in vitro,16 but the numbers of those substrates are limited. Several strategies have been proposed and conducted to globally identify kinase substrates, such as phosphoproteomics analysis after artificial trigger or reaction using anti-kinase motif antibody.21,22 One convincing strategy is global quantitative phosphoproteomics analysis using wild-type (WT)/heterozygous mutant (HET) and KO cells/tissues.23 During bioinformatics analysis, this strategy can aid in the reduction of background signals and noise.
While Nayyab et al.24 generated Tssk3 KO mice with local gene indels (−19 bp, −6+19 bp, and 47 bp) and reported similar phenotype results as our TSSK3 KO, the functional pathways and substrates of TSSK3 remain unknown. In the present report, we created Tssk3 KO mice with a deletion of 1551 bp, including most of the open reading frame (ORF), using the CRISPR/Cas9 system and revealed that TSSK3 plays an essential role in formation of sperm and male fertility. We additionally identified putative TSSK3 phosphorylation substrates using phosphoproteomics and our null mouse model. In particular, we discovered an alteration in the phosphorylation status of fertility-critical proteins in our studies.
MATERIALS AND METHODS
Ethics Statement
Mice were maintained in accordance with NIH guidelines, and all animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine.
Animals
B6D2F1 was purchased from Charles River (MA, USA). In-house hybrid mice (C57BL/6J × 129S5/SvEvBrd) were mated with Tssk3 F0 founder mouse to expand the line. For phenotypic analysis, sexually mature male mice were used. All mice were housed with a 12 h light cycle. All mouse experiments were performed according to the guidelines from the Institutional Animal Care and Use Committee at Baylor College of Medicine.
RT-PCR
mRNA was collected from tissues of C57BL6J/129SvEv hybrid mice and reverse transcribed into cDNA. Human multiple tissue cDNA was purchased from BD Biosciences. RT-PCR was performed with mouse or human cDNA as described previously25 using the following specific primers: Mouse Tssk3 (NM_080442.2) Fw, GGACTTTCTACTCTCCAATGGG, Rv, ACACAGTCAAAGACATCCCC; Mouse Hprt (NM_013556.2) Fw, TGGATATGCCCTTGACTATAATGAG, Rv, TGGCAACATCAACAGGACTC; Human TSSK3 (NM_052841.4) Fw, GTTATAGACAAGATGGGAGGGC, Rv, CATTCAGCACGCAGTCAAAG; Human GAPDH (NM_002046.7), Fw, AATCCCATCACCATCTTCCAG, Rv, ATGACCCTTTTGGCTCCC.
RNA-seq analysis
Quantitative heatmaps depicting the average transcripts per million (TPM) value per tissue per gene in a graphical output resembling conventional semi-quantitative PCR was performed as previously described.26 This information includes RNA-seq data from 6 mouse testis datasets,27 9 mouse epididymis datasets,26 10 purified mouse germ cell datasets,28,29 and 3 purified mouse Sertoli cell datasets.30 The raw values for these and other mouse and human tissues and cells can be found in Additional Files 3 and 4 (Tables S3 and S4) in our previous publication.26
Generation of Tssk3 knockout mice
Single guide RNA (sgRNA) target sequences for mouse Tssk3 were designed (AGGACTTTCTACTCTCCAAT and AGGACTTTCTACTCTCCAAT) using the CRISPRdirect suite (https://crispr.dbcls.jp/). The custom gRNAs were ordered (Sigma) and assembled into a ribonucleoprotein (RNP) complex with Cas9 protein (Thermo Fisher Scientific) at 37 ℃ for 10 min. The RNPs were electroporated into zygotes harvested from superovulated B6D2F1 females using an ECM 830 electroporation system (BTX, Holliston, MA). Embryos were cultured overnight to the 2-cell stage before being transferred into the oviducts of pseudopregnant ICR mice (Center for Comparative Medicine, Baylor College of Medicine). Founder mutations in pups born were identified by PCR and Sanger sequencing. A founder mouse with −1551 bp deletion was used to expand the colony. Mice were genotyped by PCR with specific primers for the WT alleles (Fw: 5’-AAGACTGACTTAACCCATGCTGG-3’ and Rv-WT: 5’-AGGAAAGGGCCTACCCGTGTTGG-3’) or KO alleles (Fw: 5’-AAGACTGACTTAACCCATGCTGG-3’ and Rv-KO: 5’- AGTTCCAGGAAGGTCTTTACAGG-3’).
Sperm head-tail separation
Spermatozoa from the cauda epididymis were collected into 1 ml PBS and sonicated to separate tails and heads (a Misonix Qsonica S-4000 Ultrasonic Sonicator Cell Disruptor, 20% amplitude, 10-seconds × 2 times). After the confirmation of separation through a microscope, the sample was centrifuged at 10,000 × g for 5 minutes. The sperm pellet was resuspended with 200 µl PBS, then added to 1.8 ml of 90% Percoll solution in PBS. After centrifuging at 15,000 × g for 15 minutes, the bottom layer containing heads and the top layer containing tails were separately collected. Each sample was washed with PBS three times (centrifugation at 15,000 × g for 5 min), then subjected to protein extraction.
Protein extraction and immunoblot analysis
Testes from adult male mice were homogenized using VWR Bead Mill Homogenizer in Pierce IP buffer containing protease inhibitor followed by incubation for 1 hour at 4 °C with gentle agitation. Then, the lysate was sonicated for 10 seconds with 30 second intervals at 20% amplitude and repeated 4 times. Sperm samples were resuspended in 0.5% sodium dodecyl sulfate (SDS) in PBS containing 5% 2-Mercaptoethanol. The lysate was sonicated for 10 seconds with 1 second ON and 1 second OFF at 20% amplitude. Lysates were cleared by centrifugation at 14,000 × g for 3 minutes at 4 ℃, and supernatants were subjected to SDS-PAGE under reducing conditions followed by blocking with Bullet Blocking One (Nacalai USA). The samples were then evaluated by immunoblot analysis with antibodies against TSSK3 (Proteintech, USA, 12940–1-AP) or against ZPBP1 (Cocalico Biologicals, USA, G176)31. The chemiluminescent signal was developed using ChemiDoc Imaging System (BioRad, USA).
Male fertility assessment
Sexually mature male mice were housed with two WT of C57BL6J/129SvEv hybrid females for four months. During the mating period, the number of pups born per litter per male was counted. Average litter sizes are presented as the average number of pups per litter from all the males.
Sperm count
Sperm were extracted by cutting caudal epididymis 30 times and incubating in TYH medium at 37℃ and 5% CO2. After 15 minutes incubation, sperm samples were applied to a chamber of a 100 µm-deep counting slide (CellVision), and the sperm number was measured using the Hamilton Thorne CEROS II system.
Histology
Testes and epididymides from Tssk3 KO and control mice were collected and fixed in Bouin’s fixative (Sigma Aldrich) overnight at RT and washed in 70% ethanol to remove excess stain. Tissues were embedded in paraffin, sectioned at 5 µm thickness, and stained by Periodic Acid-Schiff (PAS)-hematoxylin. Entire slides were scanned at 40X with an Aperio AT2 slide scanner (Leica Microsystems). Testis cross-sections were examined for stages of spermatogenesis by applying standard staging criteria.32
Transmission Electron Microscopy (TEM)
Testis samples were fixed in 3% glutaraldehyde and washed in 1M sodium phosphate buffer (pH 7.3). After fixing in 1% osmium tetroxide for 1 h, the samples were dehydrated. Tissue samples were hardened with acetone and Polybed 812 plastic resin and embedded in plastic molds with 100% Polybed 812. One-micron sections were cut on an ultra-Leica EMCU ultramicrotome and placed on glass slides and stained with Toluidine Blue. Ultra-thin sections (80nm) were cut from sample blocks using a Leica EMUC Ultra microtome and mounted on 100 mesh copper grids. Grids were stained with 2% uranyl acetate and Reynold’s lead stain, then analyzed on a JEOL JEM 1250 electron microscope. Images were captured on an AMTV600 digital camera at Texas Heart Institute.
Phosphoproteomics
The seminiferous tubules of testes from the adult mice were unraveled. Interstitial cells were removed by pipetting through a 1 mL syringe (Becton, Dickinson and Company, USA). Tubules were transferred to Accutase® solution (MilliporeSigma, USA), clipped into fragments with scissors, and incubated for 5 minutes at room temperature. The dissociated cell suspension was overlaid over a 70 µm mesh EASYstrainer (Greiner bio-one, USA) and centrifuged at 300 × g for 5 minutes to collect single cells. After washing with PBS, the testis-derived germ cells were harvested and subjected to the following procedure. Sample preparation for deep-scale phosphoproteomic profiling was performed as described previously,33 with minor modifications. Briefly, samples were lysed with 8 M Urea lysis buffer, reduced/alkylated with dithiothreitol/iodoacetamide and digested using LysC and trypsin enzyme. The peptides were desalted using Sep-Pak Vav 1cc C18 cartridges (Water WAT054955) and dried in a speed vac. The peptides (120 µg per sample) were labeled with TMT10plex Label Reagent Set (Thermo Scientific 90110) according to the manufacturer’s protocol. The TMT channel assignment was as follows: 5 Tssk3 HET tissues- 126, 127N, 127C, 128N, 128C; 5 KO tissues- 129N, 129C, 130N, 130C, 131. The labeled peptides were mixed and dried with a speed vacuum concentrator. Offline fractionation of TMT-labeled peptides was done using an Agilent 300Extend-C18 column (4.6 mm × 250 mm, 5 µm) on an Agilent 1260 Infinity II system at 1 ml/minute for 96 minutes. The 96 fractions were concatenated into 24 peptide pools and a flow-through pool and acidified with s final concentration of 0.1% formic acid (FA). 5% of the peptide samples were used for proteome profiling and 95% were used for phospho-proteome profiling. For the phospho-proteome, the 24 peptide pools were further concatenated to make 12 pools. The phospho-peptide enrichment was carried out using the Immobilized Metal Affinity Chromatography (IMAC) Fe-NTA agarose beads (Qiagen 1018611) as described before.33 20 µl of slurry beads per fraction tube was used to perform the enrichment and the phospho-peptides were eluted using 50% acetonitrile (ACN) containing 0.1% FA. The peptides were dried and reconstituted in loading buffer (5% methanol with 0.1% FA) for liquid chromatography tandem mass spectrometry analysis (LC-MS/MS). The peptides were separated on an online nanoflow Easy-nLC-1000 system (Thermo Fisher Scientific) coupled to Orbitrap Exploris480 mass spectrometer (Thermo Fisher Scientific). 1 µg of each fraction for proteome and 25% of each fraction for phosphoproteome was loaded on a pre-column (2 cm × 100 µm I.D.) and separated on in-line 20 cm × 75 µm I.D. column (Reprosil-Pur Basic C18aq, Dr. Maisch GmbH, Germany) equilibrated in 0.1% FA. Peptide separation was done at a flow rate of 200 nl/minute over 110 minute gradient time with different concentrations of solvent B (2% to 30% 87 minute, 30% to 60% 6 minute, 60% to 90% 7 minute, and finally hold at 50% 10 minute). The peptides were ionized at positive spray voltage (2.4 kV) and the ion transfer tube temperature was 300 ℃. The mass spectrometer was operated in a data dependent mode with 2 seconds cycle time. The MS1 was done in Orbitrap (120000 resolution, scan range 375–1500 m/z, AGC 5e5, 50 ms Injection time) followed by MS2 in Orbitrap (30000 resolution, AGC 1e5, 105 ms injection time, HCD 38%) with TurboTMT algorithm. Dynamic exclusion was set to 20 seconds and the isolation width was set to 0.7 m/z.
Phosphoproteomics Analysis and Data Processing:
Raw files were converted to mzML using MSConvert.34 Precursor ion intensities were derived from the area under each elution curve and reporter ions intensities were extracted, using MASIC.35 We used the reference mouse database (2020–03-24) provided by RefSeq.36 Philosopher37 was used to add decoy reversed sequences and common contaminants.37,38 Raw spectra were searched with MSFragger (v3.2) with mass calibration enabled.39,40 For proteomic profiling, search settings included precursor ions with charge 2 – 6, precursor mass mode set to CORRECTED, and isotope error set to −1/0/1/2. Remove precursor peak was set to true and remove precursor range was set to +/− 1.5 Da. TopN was set at 150. Fully tryptic in silico digestion settings included amino acids from 7 to 50 amino acids in length with a mass range from 350 – 10,000 Dalton generated from cleavage after KR (including if followed by P) with a maximum of 2 miscuts, and with clip N-term methionine enabled. Allow multiple variable modifications were set to false. Precursor true tolerance was set at 10 ppm and fragment mass tolerance was set at 0.02 Dalton. Fixed modifications were set for the lysine TMT label (+229.1629) and carbamidomethylation of cystine (+57.0215). Dynamic modifications were set for methionine oxidation (+15.9949), acetylation (+42.0106), peptide N-terminal TMT label (+229.1629), peptide n-term acetylation, and peptide n-term pyroGlu (−17.02650 for Q,C) and (−18.01060 for E). For phosphoprofiling, the following deviations in the search settings were made: peptide N-term acetylation was removed and a maximum of three phosphorylations per peptide on STY (79.966331) was added to the dynamic modifications list. Isotope error was set to 0/1/2 and topN peaks was set to 300. Allow modifications per peptide was set to true. Peptide validation was performed using semi-supervised learning procedure in Percolator41 as implemented in MokaPot.42 The gene product inference and iBAQ-based quantification were carried out using the gpGrouper algorithm43 to calculate peptide peak area (MS1) based expression estimates.
The median normalized and log10 transformed iBAQ values were used for data analysis. The differentially expressed proteins were calculated using the moderated t-test to calculate p-values and log2 fold changes in the R package limma. The False Discovery Rate corrected p-Value was calculated using the Benjamini-Hochberg procedure. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD038012. The motif analysis figure was generated using PhosphoSitePlus® (Cell Signaling Technology).44
Statistical Analysis
Statistical significance was evaluated using the two-tailed unpaired Student t-test assuming unequal variances except as otherwise noted. Data are represented as means ± SEM.
RESULTS
Generation of Tssk3 KO mice
We identified Tssk3 as a conserved testis-specific gene by a bioinformatic screen as previously described.26,45 We performed RT-PCR on reproductive and non-reproductive human and mouse tissues to confirm that both human TSSK3 and mouse Tssk3 are testis-specific (Fig. 1A-B). To determine the temporal expression pattern of mouse Tssk3, we performed RT-PCR using testis from WT mice of increasing age—postnatal day (PND) 5, 10, 15, 20, 25, 30, 35, and 42—to capture the earliest stage of expression of Tssk3 in mice. Expression of mouse Tssk3 was undetectable from PND 5 through PND 20, became present at a relatively low level at PND 25, increased in expression at PND 30, and remained consistent in expression at PND 35 and 42 (Fig. 1C). PND 25 corresponds to the period of spermatogenesis when round spermatids transition to elongating spermatids.46 We further utilized our previously described large-scale analysis of newly acquired and previously published RNA-seq datasets of reproductive tissues and cells to glean further insight into the testis cell populations expressing the highest levels of Tssk3 transcripts in mice. In our analysis, we included whole testes, segments of epididymis (caput, corpus, and cauda), spermatogonia [unenriched spermatogonia and ID4+ high and low spermatogonial stem cells (SSCs)], spermatocytes (leptotene and pachytene spermatocytes), round spermatids, and SOX9+ Sertoli cells.26 Our results show that round spermatids contain the highest levels of Tssk3 transcripts in mouse reproductive tissues and cells, with an expression value of 1,079±0 transcripts per million (TPM; mean±SEM) (Fig. 1D). The next highest expression was shown in whole testes with 696±65 TPM, then in Sertoli cells (116±16 TPM), pachytene spermatocytes (54.1±0 TPM), and unenriched spermatogonia (18±0 TPM) (Fig. 1D). ID4+ high and low SSCs, leptotene spermatocytes, and all three segments of the epididymis showed the lowest levels of Tssk3 expression, all with less than 6 TPM, or ~8–9-fold less than round spermatids (log2FC) (Fig. 1D).
Figure 1. Generating Tssk3 KO mice.
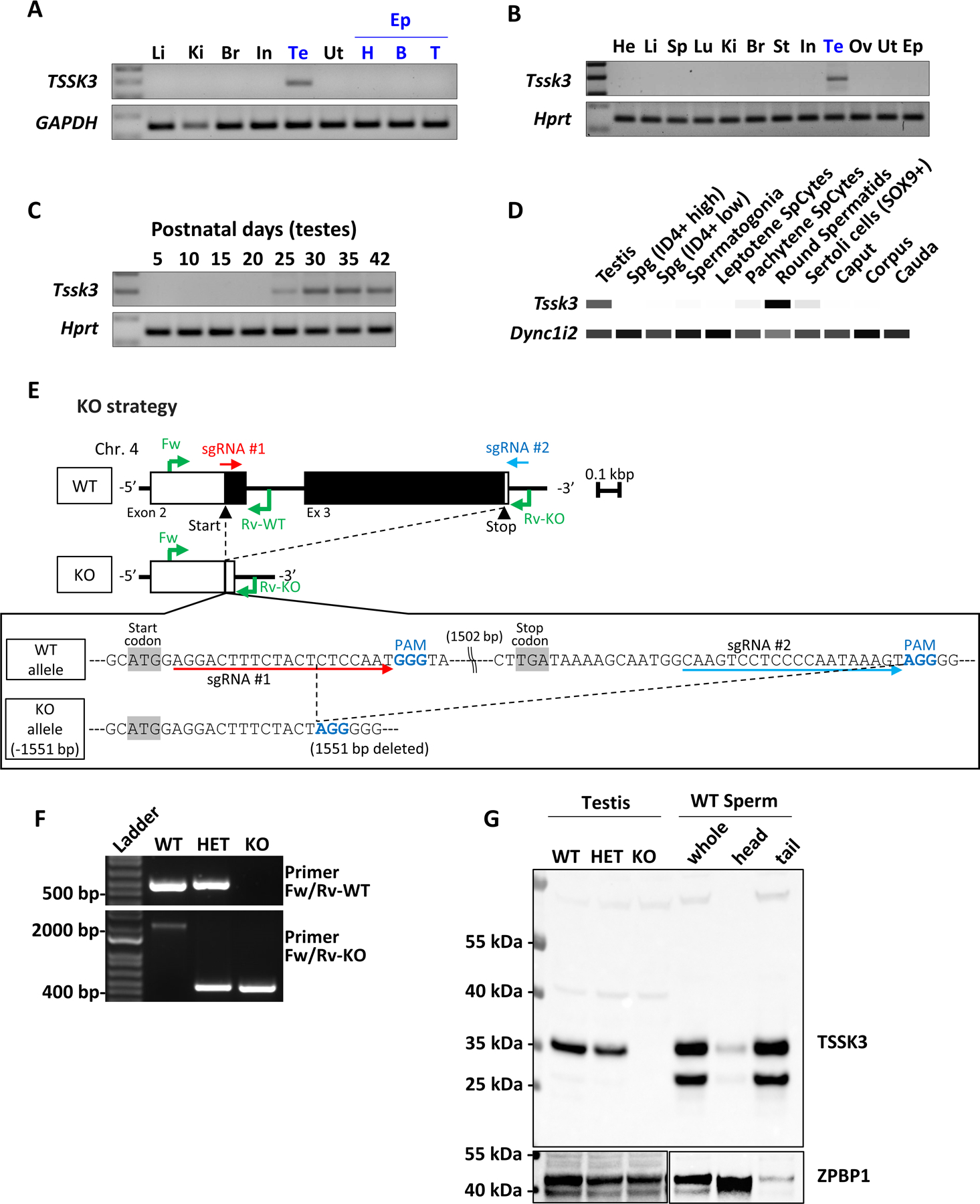
A. Multi-tissue RT-PCR expression analysis of TSSK3 in humans. GAPDH was used as a loading control. Li, Liver; Ki, Kidney; Br, Brain; In, Intestine; Te, Testis; Ut, Uterus; Ep, Epididymis; H, Head (Caput); B, Body (Corpus); T, Tail (Cauda).
B. Multi-tissue RT-PCR expression analysis of Tssk3 in mice. Hprt was used as a loading control. He, Heart; Li, Liver; Sp, Spleen; Lu, Lung; Ki, Kidney; Br, Brain; St, Stomach; In, Intestine; Te, Testis; Ov, Ovary; Ut, Uterus; Ep, Epididymis.
C. RT-PCR from mouse testes at various postnatal days.
D. Heatmap depicting RNA-seq-based Transcripts Per Million (TPM) values for mouse Tssk3 in the indicated reproductive tissues and cells. Dync1i2 was used a “loading” control. White = 0, Black = Max.
E. Genomic structure and strategy of generating Tssk3 KO mice, and the genetic sequences of mouse Tssk3 deleted by the CRISPR/Cas9 system.
F. Genotyping of Tssk3 alleles. Primers shown in Fig. 1E amplify specific amplicons for the WT or KO alleles. The sizes of several DNA ladder bands are shown for comparison.
G. Western blot analysis using testis, sperm, and spermatozoa fractionated into sperm heads and sperm tails. TSSK3 protein is detected in the testes from WT and HET, and whole sperm and sperm tails from WT mice.
To generate KO mice using the CRISPR/Cas9 system, we designed two single guide RNAs (sgRNA) for mouse Tssk3 to produce a large deletion of the coding region as shown (Fig. 1E). One-cell stage embryos were electroporated with Cas9 protein and sgRNAs, and the two-cell stage embryos were then transferred into the oviducts of pseudopregnant females. The electroporation resulted in a 1,551 bp deletion in the Tssk3 gene, corresponding to the entire region encoding the kinase domain, and almost the entire protein coding region except the first 6 amino acids of TSSK3 (Fig. 1E). After the deletion was confirmed by Sanger sequencing, specific primers for the WT or KO allele were designed and used for genotyping (Fig. 1F). The KO mice did not show any obvious developmental abnormalities or differences in sexual behavior. The absence of TSSK3 protein was confirmed by Western blot analysis using anti-TSSK3 antibodies. Interestingly, when mature WT sperm were separated into separate head and tail fractions, Western blot analysis revealed that TSSK3 is localized predominantly, if not exclusively, to the tail of mature sperm (Fig. 1G). Sperm also display a smaller form of TSSK3 of unknown significance.
Tssk3 KO males are sterile
Beginning at 6 weeks of age, sexually mature heterozygous (HET) or KO males were housed with two WT C57/129 females each for 4 months to test their fertility. The average number of offspring per litter was counted. Four control mating pairs had 9.3 ± 0.5 pups on average, whereas four homozygous mutant males had no pups and exhibited sterility (Fig. 2A). This data independently verified that TSSK3 is essential for male fertility. To determine the cause of the sterility defect, we analyzed the testis and sperm from controls and Tssk3 KO mice. The testicular size and weight of Tssk3 HET and KO mice did not show any significant difference (HET, 104.1 ± 6.4 mg versus KO, 112.0 ± 7.7 mg, Fig. 2B). Interestingly, the sperm count from caudal epididymis of KO mice was nearly ablated compared to HET males (HET, 14.26 ± 1.8 × 106/mL versus KO, 0.93 ± 0.2 × 106/mL, Fig. 2C). The few spermatozoa retrievable from the epididymis of KO males showed abnormal morphology with all KO sperm demonstrating amorphous head morphology and thin sperm midpieces (Fig. 2D).
Figure 2. Tssk3 KO causes male sterility.
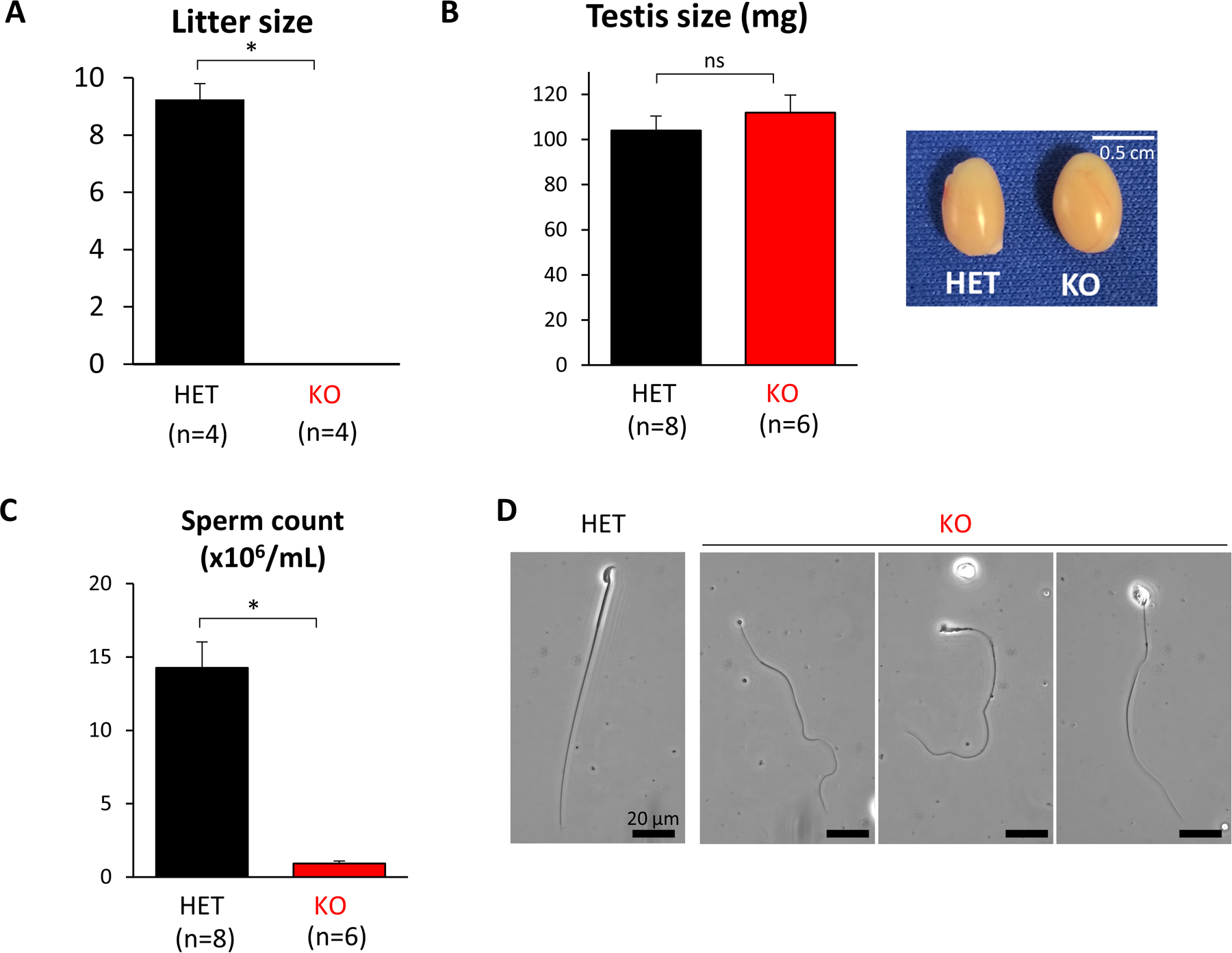
A. Average litter size from natural mating of Tssk3 HET and KO mice. Litter size was measured by the number of pups born. Tssk3 KO males showed complete infertility. P<0.00001.
B. The average weight of individual testes and images of testes from Tssk3 HET and KO mice. ns, not significant.
C. Quantification of sperm released from the cauda epididymis. Tssk3 KO males showed a significant reduction in testis volume, P<0.0001.
D. Images of Tssk3 HET and KO spermatozoa from caudal epididymis. KO sperm exhibited abnormal sperm morphology. Scale bar, 20 μm.
Microscopic analysis of control and Tssk3 KO testes, sectioned and stained with PAS-hematoxylin, revealed a mostly disorganized pattern and appearance of germ cells in the seminiferous epithelium, with all stages (I-XII) presenting some form of disorganization. In comparison to controls that have a normal appearance and organization of germ cells and easily discernable stages, most tubule cross-sections from Tssk3 KO testes presented with germ cells that appeared to be either too early or too late for that stage (Fig. 3). For example, quite commonly, elongated spermatids that should have undergone spermiation at stage VIII were observed in later stages IX-XI; early elongating spermatids that are characteristic of stage IX could be observed at XII; and, elongating spermatids that should have been fully developed at stage I were instead underdeveloped (Fig. 3). In agreement with the report by Nayyab et al.,24 we found that elongated spermatids did not line the lumen at stage VII (Fig. 3), so technically stage VII does not exist in these mice; however, it is obvious that the knockout tubules progress to stage IX since irregularly shaped round spermatids can be found, despite unreleased, fully elongated spermatids still embedded in the epithelium (Fig. 3). We also observed significant vacuolization of germ cells in histological sections of Tssk3 KO testes. Quantification of the amount of vacuolization present in KO and control testes revealed 28.7% ± 0.2% of tubule cross-sections in Tssk3 KOs contain vacuolization versus only 1.5% ± 2% in Tssk3 controls (Fig. 3 and Supplementary Fig. 1). Although the vacuolization observed in controls occurred primarily in a small percentage of degenerate tubules that naturally occur along the periphery of the testis, germ cell vacuolization as observed in the Tssk3 KO could not be found in controls. Omitting consideration of stages VII and VIII—since these two stages were nearly impossible to ascertain in Tssk3 KO mice through standard staging criteria—of all circular tubule cross-sections in the KO that contained vacuolization, 92% ± 3% were at stages IV-VI and 8% ± 3% were at stage IX (Supplementary Fig. 1). We also noted a higher distribution of stages I-VI in the KO than in controls presumably due to the obvious block or inability of elongated spermatids to proceed normally along the spermiation pathway.
Figure 3. Histological and ultrastructural analysis of testis sections from control and Tssk3 KO mice.
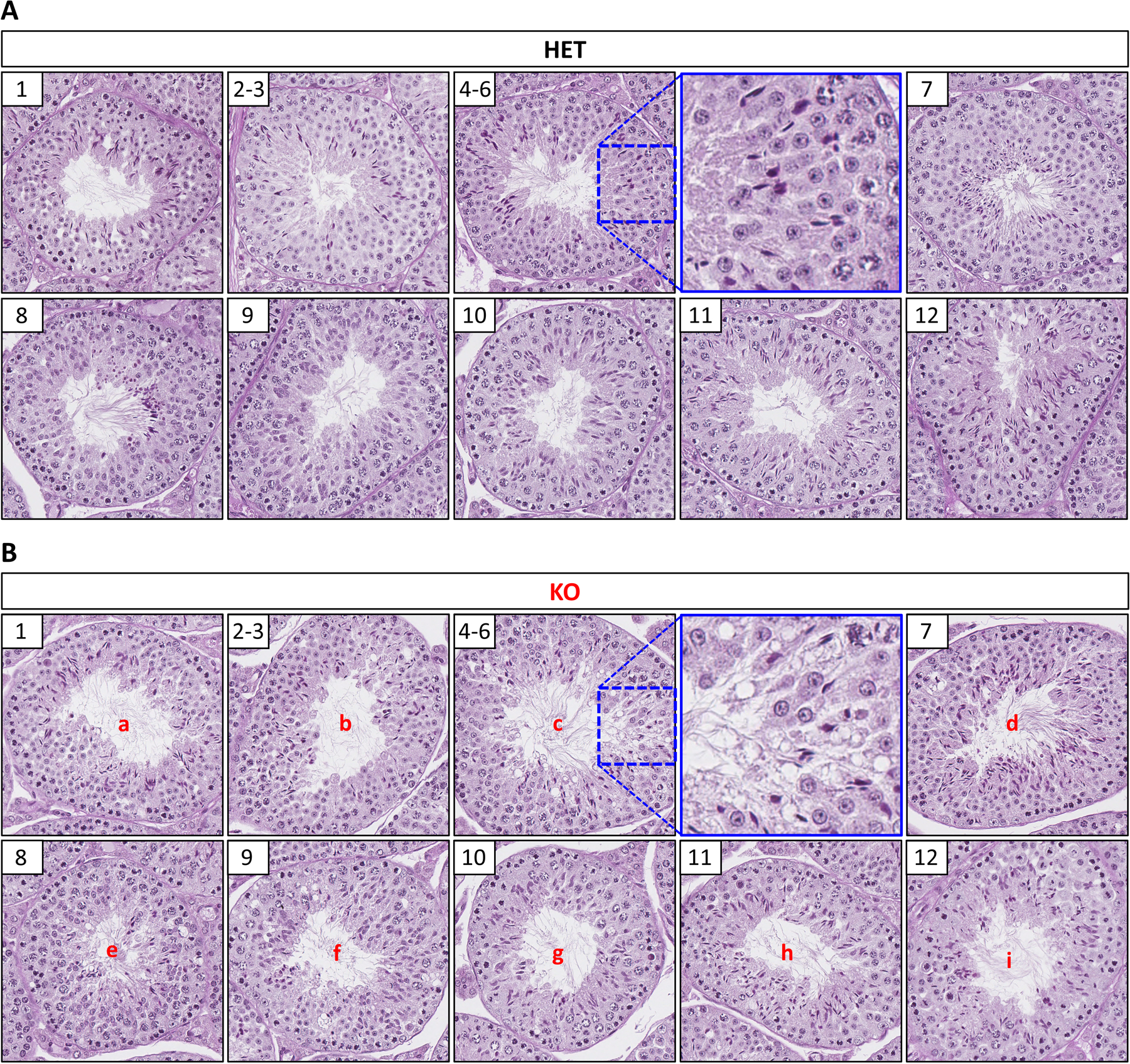
PAS-Hematoxylin staining of testis from Tssk3 HET (A) and KO (B) mice. Representative tubule cross-sections from each stage (I-XII) as indicated in the white box in the upper left-hand corner of each image. Higher magnification insets for Stage IV-VI are included as indicated with blue boxes to better show vacuolization present in KO (B), but not HET (A) mice. Abnormalities observed in the Tssk3 KO (B) mice are indicated as follows: a, elongated spermatids not fully condensed; b, disorganized appearance of elongated spermatids; c, significant vacuolization present; d, elongated spermatids embedded in epithelium (not lining the lumen), vacuolization; e, unreleased elongated spermatids, vacuolization; f, a mixture of early elongating spermatids and unreleased fully elongated spermatids, vacuolization; g, a mixture of early elongating spermatids and unreleased fully elongated spermatids; h, a mixture of developing (not fully condensed) elongated spermatids and unreleased fully elongated spermatids; i, a mixture of developing (not fully condensed) elongated spermatids and unreleased fully elongated spermatids. Scale bar, 50 μm.
Analysis of epididymis histology revealed that only a few sperm were present in the lumen of KO mice, while, as expected, the lumen of WT mice was filled with sperm (Fig. 4A), which correlates with our sperm count results (Fig. 2C). We further investigated the morphology of sperm in the testes by transmission electron microscopy (TEM). Similar to our light microscopy observations, we found abnormalities present through TEM. Although acrosomal shape and the manchette in KO elongated spermatids at step 9 appeared normal, at steps 14 and 15, they showed unstructured morphology and degradation, while condensation of nuclei was predominantly observed (Fig. 4B). Notably, TEM analysis revealed the detachment of the acrosomes from the nuclei of Tssk3 KO sperm, which has been previously observed in mice containing deletions of genes coding actin-related proteins (ARPs), such as actin-like 7a (Actl7a) and actin-like 9 (Actl9).47,48 Taken together, our findings indicate that oligoteratozoospermia in Tssk3 KO males results from defective spermiogenesis and spermiation that leads to degeneration and reabsorption of nearly the entire population of mature spermatozoa.
Figure 4. Histological of epididymis sections and ultrastructural analysis of testis sections from control and Tssk3 KO mice.
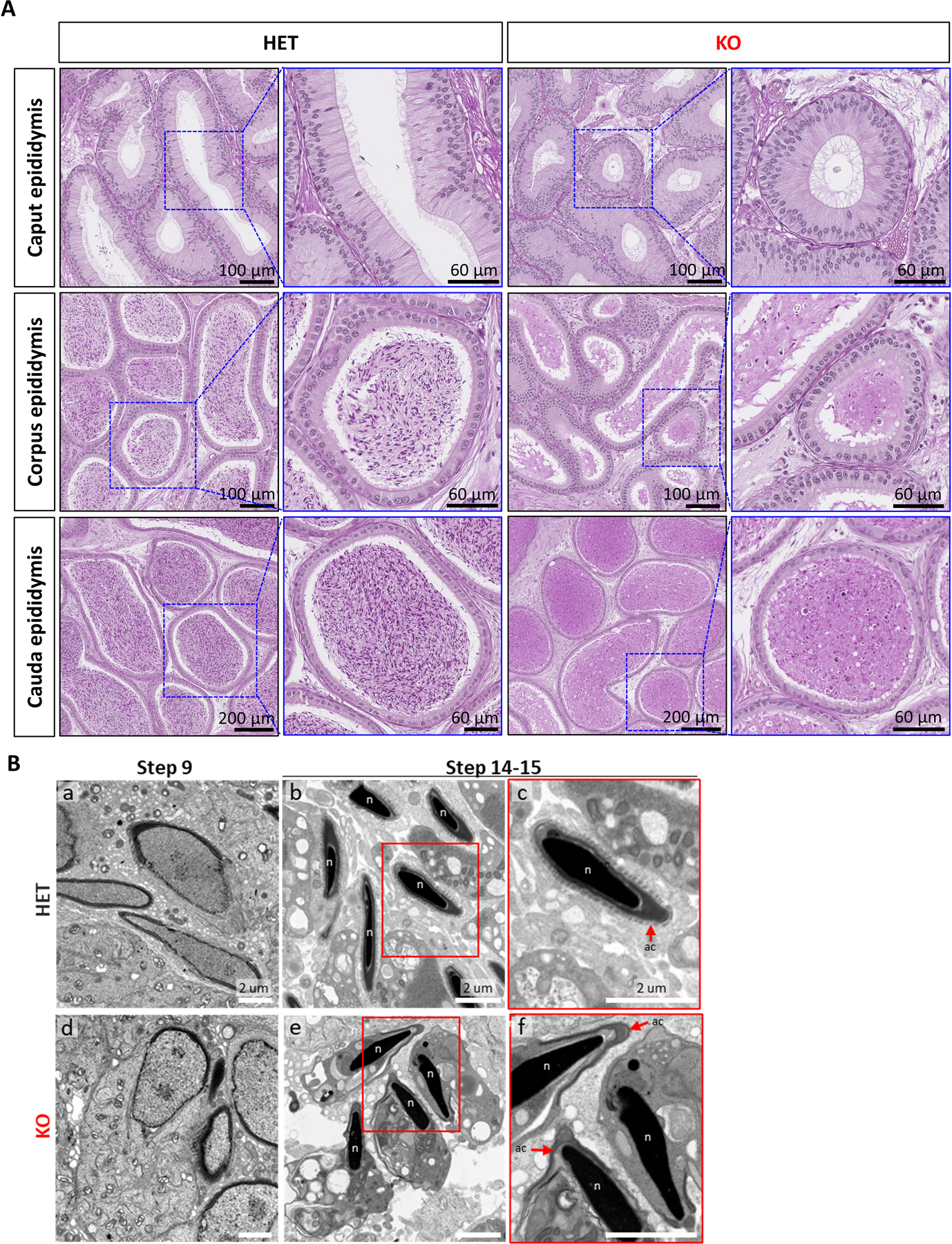
A. PAS-Hematoxylin staining of epididymis from Tssk3 HET and KO mice. Scale bars as indicated.
B. Transmission electron microscopy images of Tssk3 HET and KO testis. (a) HET and (d) KO spermatids at step 9 (Scale bar, 2 μm). Spermatozoa at step 14–15 from (b) HET and (e) KO (Scale bar, 2 μm). Tssk3 KO sperm have cytoplasm around normal-shaped nuclei (n). The magnified images, (c) and (f) show perspectives in white squares on (b) and (f), respectively. KO sperm have detached acrosome (ac, red arrows) from the sperm nuclei (n) (Scale bar, 2 μm).
Phosphoproteomics
To investigate TSSK3 substrates in vivo, we conducted quantitative phosphoproteomic profiling of testis germ cells from Tssk3 HET and KO mice (n=5 mice/genotype). Using isobaric-based TMT labeling, these samples were subjected to the mass spectrometer for quantification profiling and quantitative phosphoproteomics. We obtained distinct distribution patterns between HET and KO groups in the first principal component analysis in both expression profiling and phosphoproteomics (Supplementary Data). In total, we acquired 8,430 proteins, 3,908 phosphorylated proteins, and 22,399 phosphopeptides from Tssk3 HET and KO testes (Supplementary Data). To generate our selective list of TSSK3-altered proteins and phosphoproteins, we set our data-filtration parameters to a fold-change > 1.25 and P-value < 0.05. Consequently, we found 335 proteins were differently expressed [135 up-regulated (blue dots) and 200 down-regulated (red dots)] in HET testes compared to KO testes (Fig. 5A). In terms of phosphorylation, 276 proteins were differently phosphorylated [167 proteins highly-phosphorylated (blue dots) and 109 phosphorylated at lower-level (red dots)] in HET testes compared to KO testes (Fig. 5B). To investigate potential TSSK3 phosphosites of substrates, we selected the top 100 upregulated phosphosite in HET and subjected them to PhosphositePlus (https://www.phosphosite.org/sequenceLogoAction) for multiple sequence alignment (Fig. 5C) and MotifFinder on MOTIF Search (https://www.genome.jp/tools/motif/). Previously, the RRSSSV(Y) motif was reported as a TSSK3 substrate in vitro.49 In our present in vivo data, we did not find this motif as highly phosphorylated. To further explore the implications of our data, we manually curated proteins within our lists that have been previously reported to be critical for male fertility. In Fig. 5D, we present top fertility-related proteins that are either highly-expressed in HET (downregulated in KO), highly-phosphorylated in HET (decreased in phosphorylation in KO, that may or may not be due to a change in total protein level), or highly-phosphorylated in HET with unchanged total protein levels in HET (truly decreased in phosphorylation in KO). In the highly-expressed group, which could be due to the apparent degradation and reabsorption of sperm in the testes, OAZ3, TPPP2, GAPDHS, PGK2, and TCP11 were found (Fig. 5D). In our highly-phosphorylated list, we discovered ACTL7A, FAM71D, GAPDHS, SMCP, SPATA19, and TPPP2; and, in our highly-phosphorylated list with unchanged protein expression, we found UBE3B, CFAP57, ACTL9, and REEP6. These data strongly suggest that TSSK3 works as a crucial kinase to directly or indirectly phosphorylate multiple proteins responsible for spermatogenesis and sperm function.
Figure 5. Phosphoproteomic analysis to uncover putative TSSK3 substrates.
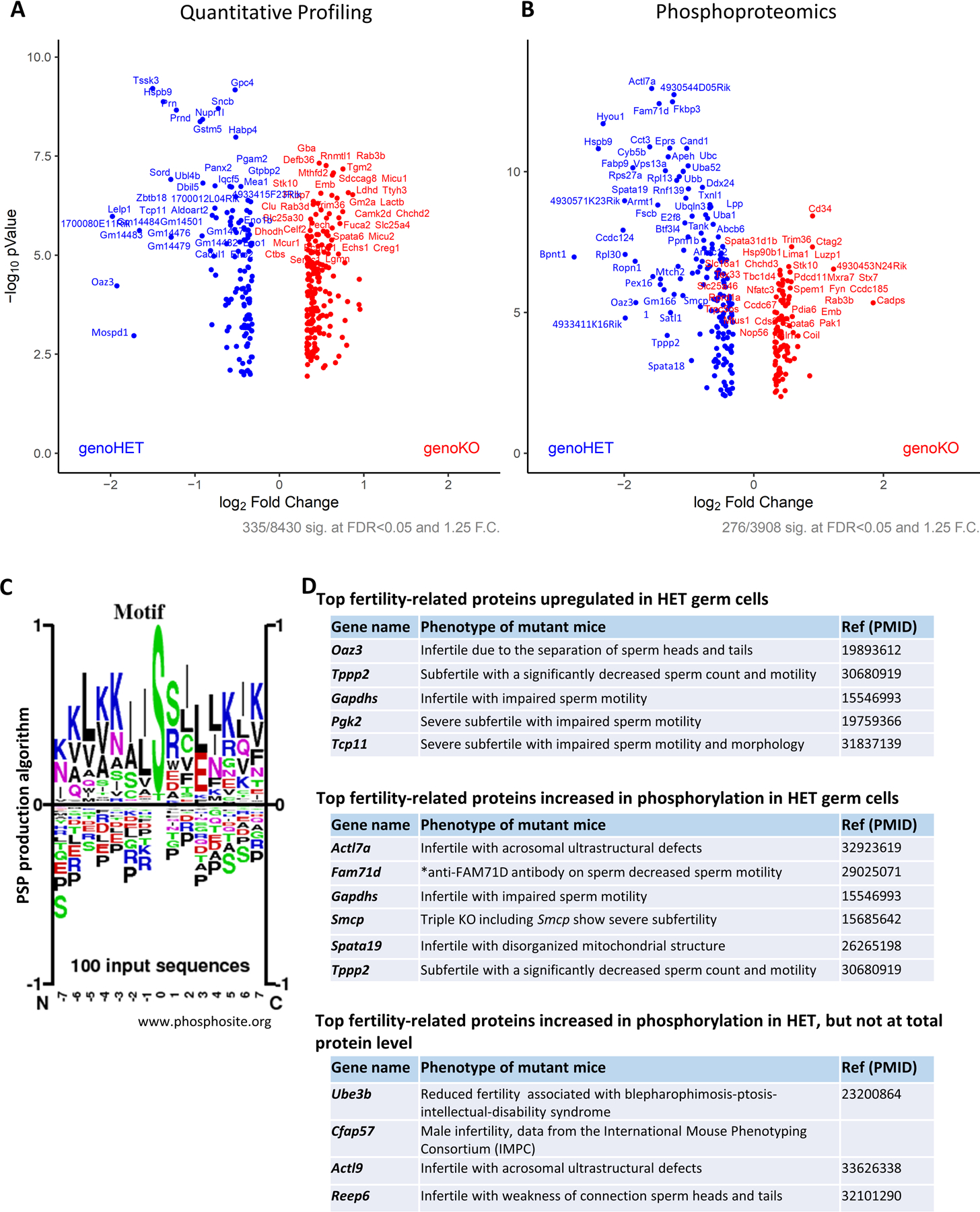
A. Volcano plot of quantitative analysis of proteins identifies MS profiling. Proteins expressed with fold-changes (HET: KO ratio) > 1.25. (p < 0.05) were selected.
B. Volcano plot of quantitative analysis of phosphopeptides identified by MS. Phospho-proteins with fold-changes (HET: KO ratio) > 1.25. (p < 0.05) were selected.
C. Logo motif of the phosphosites. The top 100 phosphosites phosphorylated at higher levels in Tssk3 HET than in KO testes were subjected to the sequence logos creation using PhosphoSitePlus®.
D. The lists of upregulated or phosphorylated proteins in HET compared to KO, that are reported in each mutant mouse model show infertility/subfertility phenotype.
DISCUSSION
Herein, by generating a Tssk3 KO mouse model and demonstrating an altered proteome and phosphoproteome, we have demonstrated that TSSK3 is an essential kinase for male fertility. Tssk3 KO mice are sterile due to abnormal drastic reduction in sperm production and abnormal sperm morphology (oligoteratozoospermia). The abnormalities in spermatogenesis due to the absence of TSSK3 did not show an obvious change until late spermatogenesis in histological and ultrastructural analysis (Fig 3A-B, 4B). This observation and expression analysis of the mouse Tssk3 gene (Fig. 1A-D) suggests that TSSK3 starts to function during or after the elongation stage in spermiogenesis, where spermatozoa dynamically transform sperm shape. Our phosphoproteomics findings implicate multiple proteins as substrates of TSSK3. OAZ3 and REEP6 have been reported to be essential for a rigid junction between the sperm head and tail, critical for proper sperm shape.50,51 The KO mice of Tppp2 or Tcp11 result in impaired sperm production.52,53 TPPP2 and SPATA19 were reported to be critical for the mitochondrial function in the sperm midpiece.52,54 FAM71D and SMCP have been suggested to function in sperm motility.55,56 GAPDHS and PGK2 are known not to affect spermatogenesis but have important functions in regulating sperm motility through metabolism.57,58 Moreover, sperm from ACTL7A and ACTL9 KO male mice have been previously reported to show detachment of the acrosome from the nuclei, which we observed in Tssk3 KO sperm.47,48 Our data that shows localization of TSSK3 to the tail suggests that it interacts with and/or results in the phosphorylation of the cytoskeleton,59 as we observed significant changes in the phosphorylation state of ACTL7A and ACTL9, which are “actin like” constituents of the cytoskeleton (Fig. 5D). The interaction of TSSK3 and cytoskeletal proteins was also suggested in the previous report.24 However, because Tssk3 KO males do not phenocopy Actl7a and Actl9 KO males that only show a reduction in sperm counts, and had more severe phenotypes,47,48 other proteins (presented above) must also function downstream of TSSK3. Additionally, as we observed a widely different pattern of phosphorylation in testis germ cells between Tssk3 HET and KO in our phosphoproteomic analysis, TSSK3 phosphorylates or induces phosphorylation of multiple proteins in addition to ACTL7A and ACTL9. Further studies will be required in the future to verify how other TSSK3-induced phosphorylated proteins are involved in the male sterility phenotype of Tssk3 KO mice. The question of whether the phosphorylation of these proteins occurs directly or indirectly by TSSK3 would be interesting as well.
To date, TSSK3 has been relatively unexplored in terms of its substrates. Only a potential phosphosite has been postulated through an in vitro assay.49 In our global phosphoproteomics study, we found that TSSK3 phosphorylates, or at least participates in the phosphorylation of a large number of proteins. Interestingly, GAPDHS showed upregulation in both quantitative profiling and phosphorylation (Fig. 5D). KO male mice of Gapdhs do not show abnormalities in spermatogenesis but have abnormalities in metabolism related to regulation of sperm motility, which in turn results in male infertility.57 Since Tssk3 KO mice in this study displayed degeneration and rapid absorption of elongating/elongated spermatids in testes, normal sperm at the cauda epididymis could not be obtained. Thus, although we confirmed the importance of TSSK3 for proper sperm development during spermiogenesis, the function of TSSK3 in mature spermatozoa could not be examined. Our findings show that TSSK3 is predominantly localized to the tail of mature spermatozoa in WT (Fig. 1G). Whether TSSK3 regulates GAPDHS in mature spermatozoa remains to be determined. Because Tssk3 is expressed predominantly in spermatids (Fig. 1D), we speculate that the bulk of the phenotypic response and proteomic/phosphoproteomic results come from spermatids. The testicular germ cell enrichment strategy that we used to perform our proteomic and phosphoproteomic analyses depleted interstitial cells but did not enrich for spermatids or deplete Sertoli cells; however, because germ cell-specific proteins were mainly altered in phosphoproteomic analysis, our knockout phenotype is cell autonomous.
Our data is the first report to approach TSSK3 substrates in vivo using Tssk3 KO mice and a global proteomics analysis approach, which is an unavoidable and necessary step to identifying kinase substrates to understand the function(s) of a protein kinase. We here show that generating KO mice using the CRISPR/Cas9 system is a powerful gateway to exploring the function of causative genes for male infertility in vivo. We may, however, have overlooked other TSSK3 substrates which were not trapped in our procedure. For example, we did not recognize the phosphosite previously found for TSSK3 through in vitro studies.49 To conclude identifying kinase substrates, we may need to compare phenotypic or biological test data with proteomics data. Future studies will investigate the potential relevance of TSSK3 and infertile patients as well. Polymorphisms in human TSSK3 have been reported in the dbSNP database (www.ncbi.nlm.nih.gov/snp), suggesting that this gene could be a cause of idiopathic infertility in men displaying oligoteratozoospermia. Simultaneously, TSSK3 and its substrates could be strong candidates as male contraceptive targets. The findings presented here hold promise for new knowledge on the diagnosis and management of male infertility as well as the development of non-hormonal contraceptives.
CONCLUSION
In conclusion, our studies using Tssk3 KO mice and proteomics have revealed that TSSK3 is essential for male fertility and is a critical kinase involved in the phosphorylation of multiple infertility-related proteins. These findings provide novel insights into the treatment of the infertile male and identifying a vulnerable target for non-hormonal male contraception.
Supplementary Material
Supplementary Figure 1
Sixty individually cropped PAS/H-stained seminiferous tubule cross-sections displaying the vacuolization abnormality widespread throughout Tssk3 KO mouse testes. This vacuolization is not present in controls. All panels at same magnification. Scale bar, 50 μm.
Supplementary Table 1.
Dataset of the quantitative profiling and the phosphoproteomics data.
Supplementary Table 2.
Differential phosphosites lists.
ACKNOWLEDGMENTS
This study has been supported by the Bill and Melinda Gates Foundation (INV-001902; to M.M.M., M.I., and T.X.G), National Institutes of Health R01 HD106056 and R01 HD095341 (to T.X.G.) and R01 HD088412 (to M.M.M. and T.X.G.), a Japan Society for the Promotion of Science Overseas Research Fellowship (to K.N.), and the Lalor Foundation (to K.N.). The BCM Mass Spectrometry Proteomics Core is supported by the Dan L. Duncan Comprehensive Cancer Center NIH award (P30 CA125123), CPRIT Core Facility Award (RP210227), Intellectual Development Disabilities Research Center Award (P50 HD103555), and an NIH High End Instrument Award (S10 OD026804). We thank Maria Singhal and Benito Carbajal Jr. with Community Pathology Associates at Baylor College of Medicine for histology services, Thomas Huynh with the Department of Veterinary Medicine and Surgery at the University of Texas MD Anderson Cancer Center for Aperio AT2 slide scanner access, and Dr. Haruhiko Miyata at Osaka University for experimental advice.
REFERENCES
- 1.Berookhim BM, Schlegel PN. Azoospermia due to spermatogenic failure. Urol Clin North Am. 2014; 41(1): 97–113. 10.1016/j.ucl.2013.08.004 [DOI] [PubMed] [Google Scholar]
- 2.Oud MS, Smits RM, Smith HE, Mastrorosa FK, Holt GS, Houston BJ, de Vries PF, Alobaidi BKS, Batty LE, Ismail H, Greenwood J, Sheth H, Mikulasova A, Astuti GDN, Gilissen C, McEleny K, Turner H, Coxhead J, Cockell S, Braat DDM, Fleischer K, D’Hauwers KWM, Schaafsma E, Genetics of Male Infertility Initiative c, Nagirnaja L, Conrad DF, Friedrich C, Kliesch S, Aston KI, A Riera-Escamilla, C Krausz, Gonzaga-Jauregui C, Santibanez-Koref M, Elliott DJ, Vissers L, Tuttelmann F, O’Bryan MK, Ramos L, Xavier MJ, van der Heijden GW, Veltman JA. A de novo paradigm for male infertility. Nat Commun 2022; 13(1): 154. 10.1038/s41467-021-27132-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houston BJ, Riera-Escamilla A, Wyrwoll MJ, Salas-Huetos A, Xavier MJ, Nagirnaja L, Friedrich C, Conrad DF, Aston KI, Krausz C, Tuttelmann F, O’Bryan MK, Veltman JA, Oud MS. A systematic review of the validated monogenic causes of human male infertility: 2020 update and a discussion of emerging gene-disease relationships. Hum Reprod Update 2021; 28(1): 15–29. 10.1093/humupd/dmab030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel B, Parets S, Akana M, Kellogg G, Jansen M, Chang C, Cai Y, Fox R, Niknazar M, Shraga R, Hunter C, Pollock A, Wisotzkey R, Jaremko M, Bisignano A, Puig O. Comprehensive genetic testing for female and male infertility using next-generation sequencing. J Assist Reprod Genet 2018; 35(8): 1489–1496. 10.1007/s10815-018-1204-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oud MS, Volozonoka L, Smits RM, Vissers L, Ramos L, Veltman JA. A systematic review and standardized clinical validity assessment of male infertility genes. Hum Reprod 2019; 34(5): 932–941. 10.1093/humrep/dez022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kent K, Johnston M, Strump N, Garcia TX. Toward Development of the Male Pill: A Decade of Potential Non-hormonal Contraceptive Targets. Front Cell Dev Biol 2020; 8:61. 10.3389/fcell.2020.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nozawa K, Zhang Q, Miyata H, Devlin DJ, Yu Z, Oura S, Koyano T, Matsuyama M, Ikawa M, Matzuk MM. Knockout of serine-rich single-pass membrane protein 1 (Ssmem1) causes globozoospermia and sterility in male mice. Biol Reprod 2020; 103(2): 244–253. 10.1093/biolre/ioaa040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griswold MD. Spermatogenesis: The Commitment to Meiosis. Physiol Rev 2016; 96(1): 1–17. 10.1152/physrev.00013.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zakrzewski P, Lenartowski R, Redowicz MJ, Miller KG, Lenartowska M. Expression and localization of myosin VI in developing mouse spermatids. Histochem Cell Biol 2017; 148(4): 445–462. 10.1007/s00418-017-1579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sefton BM. Overview of protein phosphorylation. Curr Protoc Cell Biol 2001; Chapter 14(Unit 14 11. 10.1002/0471143030.cb1401s00 [DOI] [PubMed]
- 11.Moss SB, Turner RM, Burkert KL, VanScoy Butt H, Gerton GL. Conservation and function of a bovine sperm A-kinase anchor protein homologous to mouse AKAP82. Biol Reprod 1999; 61(2): 335–342. 10.1095/biolreprod61.2.335 [DOI] [PubMed] [Google Scholar]
- 12.Pereira R, Sa R, Barros A, Sousa M. Major regulatory mechanisms involved in sperm motility. Asian J Androl 2017; 19(1): 5–14. 10.4103/1008-682X.167716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nolan MA, Babcock DF, Wennemuth G, Brown W, Burton KA, McKnight GS. Sperm-specific protein kinase A catalytic subunit Calpha2 orchestrates cAMP signaling for male fertility. Proc Natl Acad Sci U S A 2004; 101(37): 13483–13488. 10.1073/pnas.0405580101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma H, Zhang B, Khan A, Zhao D, Ma A, Zhou J, Khan I, Khan K, Zhang H, Zhang Y, Jiang X, Dil S, Zeb A, Rahim F, Shi Q. Novel frameshift mutation in STK33 is associated with asthenozoospermia and multiple morphological abnormalities of the flagella. Hum Mol Genet 2021; 30(21): 1977–1984. 10.1093/hmg/ddab165 [DOI] [PubMed] [Google Scholar]
- 15.Kawa S, Ito C, Toyama Y, Maekawa M, Tezuka T, Nakamura T, Nakazawa T, Yokoyama K, Yoshida N, Toshimori K, Yamamoto T. Azoospermia in mice with targeted disruption of the Brek/Lmtk2 (brain-enriched kinase/lemur tyrosine kinase 2) gene. Proc Natl Acad Sci U S A 2006; 103(51): 19344–19349. 10.1073/pnas.0603603103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salicioni AM, Gervasi MG, Sosnik J, Tourzani DA, Nayyab S, Caraballo DA, Visconti PE. Testis-specific serine kinase protein family in male fertility and as targets for non-hormonal male contraception. Biol Reprod 2020; 103(2): 264–274. 10.1093/biolre/ioaa064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martins LR, Bung RK, Koch S, Richter K, Schwarzmuller L, Terhardt D, Kurtulmus B, Niehrs C, Rouhi A, Lohmann I, Pereira G, Frohling S, Glimm H, Scholl C. Stk33 is required for spermatid differentiation and male fertility in mice. Dev Biol 2018; 433(1): 84–93. 10.1016/j.ydbio.2017.11.007 [DOI] [PubMed] [Google Scholar]
- 18.Shang P, Baarends WM, Hoogerbrugge J, Ooms MP, van Cappellen WA, de Jong AA, Dohle GR, van Eenennaam H, Gossen JA, Grootegoed JA. Functional transformation of the chromatoid body in mouse spermatids requires testis-specific serine/threonine kinases. J Cell Sci 2010; 123(Pt 3): 331–339. 10.1242/jcs.059949 [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Wei Y, Fu G, Li H, Saiyin H, Lin G, Wang Z, Chen S, Yu L. Tssk4 is essential for maintaining the structural integrity of sperm flagellum. Mol Hum Reprod 2015; 21(2): 136–145. 10.1093/molehr/gau097 [DOI] [PubMed] [Google Scholar]
- 20.Spiridonov NA, Wong L, Zerfas PM, Starost MF, Pack SD, Paweletz CP, Johnson GR. Identification and characterization of SSTK, a serine/threonine protein kinase essential for male fertility. Mol Cell Biol 2005; 25(10): 4250–4261. 10.1128/MCB.25.10.4250-4261.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman NJ, Parker BL, Chaudhuri R, Fisher-Wellman KH, Kleinert M, Humphrey SJ, Yang P, Holliday M, Trefely S, Fazakerley DJ, Stockli J, Burchfield JG, Jensen TE, Jothi R, Kiens B, Wojtaszewski JF, Richter EA, James DE. Global Phosphoproteomic Analysis of Human Skeletal Muscle Reveals a Network of Exercise-Regulated Kinases and AMPK Substrates. Cell Metab 2015; 22(5): 922–935. 10.1016/j.cmet.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ducommun S, Deak M, Sumpton D, Ford RJ, Nunez Galindo A, Kussmann M, Viollet B, Steinberg GR, Foretz M, Dayon L, Morrice NA, Sakamoto K. Motif affinity and mass spectrometry proteomic approach for the discovery of cellular AMPK targets: identification of mitochondrial fission factor as a new AMPK substrate. Cell Signal 2015; 27(5): 978–988. 10.1016/j.cellsig.2015.02.008 [DOI] [PubMed] [Google Scholar]
- 23.Chen Z, Lei C, Wang C, Li N, Srivastava M, Tang M, Zhang H, Choi JM, Jung SY, Qin J, Chen J. Global phosphoproteomic analysis reveals ARMC10 as an AMPK substrate that regulates mitochondrial dynamics. Nat Commun 2019; 10(1): 104. 10.1038/s41467-018-08004-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nayyab S, Gervasi MG, Tourzani DA, Caraballo DA, Jha KN, Teves ME, Cui W, Georg GI, Visconti PE, Salicioni AM. TSSK3, a novel target for male contraception, is required for spermiogenesis. Mol Reprod Dev 2021; 88(11): 718–730. 10.1002/mrd.23539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holcomb RJ, Oura S, Nozawa K, Kent K, Yu Z, Robertson MJ, Coarfa C, Matzuk MM, Ikawa M, Garcia TX. The testis-specific serine proteases PRSS44, PRSS46, and PRSS54 are dispensable for male mouse fertility. Biol Reprod 2020; 102(1): 84–91. 10.1093/biolre/ioz158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson MJ, Kent K, Tharp N, Nozawa K, Dean L, Mathew M, Grimm SL, Yu Z, Legare C, Fujihara Y, Ikawa M, Sullivan R, Coarfa C, Matzuk MM, Garcia TX. Large-scale discovery of male reproductive tract-specific genes through analysis of RNA-seq datasets. BMC Biol 2020; 18(1): 103. 10.1186/s12915-020-00826-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489(7414): 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helsel AR, Yang QE, Oatley MJ, Lord T, Sablitzky F, Oatley JM. ID4 levels dictate the stem cell state in mouse spermatogonia. Development 2017; 144(4): 624–634. 10.1242/dev.146928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.da Cruz I, Rodriguez-Casuriaga R, Santinaque FF, Farias J, Curti G, Capoano CA, Folle GA, Benavente R, Sotelo-Silveira JR, Geisinger A. Transcriptome analysis of highly purified mouse spermatogenic cell populations: gene expression signatures switch from meiotic-to postmeiotic-related processes at pachytene stage. BMC Genomics 2016; 17(294. 10.1186/s12864-016-2618-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmermann C, Stevant I, Borel C, Conne B, Pitetti JL, Calvel P, Kaessmann H, Jegou B, Chalmel F, Nef S. Research resource: the dynamic transcriptional profile of sertoli cells during the progression of spermatogenesis. Mol Endocrinol 2015; 29(4): 627–642. 10.1210/me.2014-1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin YN, Roy A, Yan W, Burns KH, Matzuk MM. Loss of zona pellucida binding proteins in the acrosomal matrix disrupts acrosome biogenesis and sperm morphogenesis. Mol Cell Biol 2007; 27(19): 6794–6805. 10.1128/MCB.01029-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meistrich ML, Hess RA. Assessment of spermatogenesis through staging of seminiferous tubules. Methods Mol Biol 2013; 927(299–307. 10.1007/978-1-62703-038-0_27 [DOI] [PubMed] [Google Scholar]
- 33.Mertins P, Tang LC, Krug K, Clark DJ, Gritsenko MA, Chen L, Clauser KR, Clauss TR, Shah P, Gillette MA, Petyuk VA, Thomas SN, Mani DR, Mundt F, Moore RJ, Hu Y, Zhao R, Schnaubelt M, Keshishian H, Monroe ME, Zhang Z, Udeshi ND, Mani D, Davies SR, Townsend RR, Chan DW, Smith RD, Zhang H, Liu T, Carr SA. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat Protoc 2018; 13(7): 1632–1661. 10.1038/s41596-018-0006-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak MY, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 2012; 30(10): 918–920. 10.1038/nbt.2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monroe ME, Shaw JL, Daly DS, Adkins JN, Smith RD. MASIC: a software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Comput Biol Chem 2008; 32(3): 215–217. 10.1016/j.compbiolchem.2008.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 2016; 44(D1): D733–745. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.da Veiga Leprevost F, Haynes SE, Avtonomov DM, Chang HY, Shanmugam AK, Mellacheruvu D, Kong AT, Nesvizhskii AI. Philosopher: a versatile toolkit for shotgun proteomics data analysis. Nat Methods 2020; 17(9): 869–870. 10.1038/s41592-020-0912-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, Halim VA, Bagshaw RD, Hubner NC, Al-Hakim A, Bouchard A, Faubert D, Fermin D, Dunham WH, Goudreault M, Lin ZY, Badillo BG, Pawson T, Durocher D, Coulombe B, Aebersold R, Superti-Furga G, Colinge J, Heck AJ, Choi H, Gstaiger M, Mohammed S, Cristea IM, Bennett KL, Washburn MP, Raught B, Ewing RM, Gingras AC, Nesvizhskii AI. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods 2013; 10(8): 730–736. 10.1038/nmeth.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D, Nesvizhskii AI. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat Methods 2017; 14(5): 513–520. 10.1038/nmeth.4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu F, Teo GC, Kong AT, Haynes SE, Avtonomov DM, Geiszler DJ, Nesvizhskii AI. Identification of modified peptides using localization-aware open search. Nat Commun 2020; 11(1): 4065. 10.1038/s41467-020-17921-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson DC, Li W, Payan DG, Noble WS. A new algorithm for the evaluation of shotgun peptide sequencing in proteomics: support vector machine classification of peptide MS/MS spectra and SEQUEST scores. J Proteome Res 2003; 2(2): 137–146. 10.1021/pr0255654 [DOI] [PubMed] [Google Scholar]
- 42.Fondrie WE, Noble WS. mokapot: Fast and Flexible Semisupervised Learning for Peptide Detection. J Proteome Res 2021; 20(4): 1966–1971. 10.1021/acs.jproteome.0c01010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saltzman AB, Leng M, Bhatt B, Singh P, Chan DW, Dobrolecki L, Chandrasekaran H, Choi JM, Jain A, Jung SY, Lewis MT, Ellis MJ, Malovannaya A. gpGrouper: A Peptide Grouping Algorithm for Gene-Centric Inference and Quantitation of Bottom-Up Proteomics Data. Mol Cell Proteomics 2018; 17(11): 2270–2283. 10.1074/mcp.TIR118.000850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res 2012; 40(Database issue): D261–270. 10.1093/nar/gkr1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyata H, Castaneda JM, Fujihara Y, Yu Z, Archambeault DR, Isotani A, Kiyozumi D, Kriseman ML, Mashiko D, Matsumura T, Matzuk RM, Mori M, Noda T, Oji A, Okabe M, Prunskaite-Hyyrylainen R, Ramirez-Solis R, Satouh Y, Zhang Q, Ikawa M, Matzuk MM. Genome engineering uncovers 54 evolutionarily conserved and testis-enriched genes that are not required for male fertility in mice. Proc Natl Acad Sci U S A 2016; 113(28): 7704–7710. 10.1073/pnas.1608458113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bellve AR, Cavicchia JC, Millette CF, O’Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol 1977; 74(1): 68–85. 10.1083/jcb.74.1.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xin A, Qu R, Chen G, Zhang L, Chen J, Tao C, Fu J, Tang J, Ru Y, Chen Y, Peng X, Shi H, Zhang F, Sun X. Disruption in ACTL7A causes acrosomal ultrastructural defects in human and mouse sperm as a novel male factor inducing early embryonic arrest. Sci Adv 2020; 6(35): eaaz4796. 10.1126/sciadv.aaz4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dai J, Zhang T, Guo J, Zhou Q, Gu Y, Zhang J, Hu L, Zong Y, Song J, Zhang S, Dai C, Gong F, Lu G, Zheng W, Lin G. Homozygous pathogenic variants in ACTL9 cause fertilization failure and male infertility in humans and mice. Am J Hum Genet 2021; 108(3): 469–481. 10.1016/j.ajhg.2021.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bucko-Justyna M, Lipinski L, Burgering BM, Trzeciak L. Characterization of testis-specific serine-threonine kinase 3 and its activation by phosphoinositide-dependent kinase-1-dependent signalling. FEBS J 2005; 272(24): 6310–6323. 10.1111/j.1742-4658.2005.05018.x [DOI] [PubMed] [Google Scholar]
- 50.Tokuhiro K, Isotani A, Yokota S, Yano Y, Oshio S, Hirose M, Wada M, Fujita K, Ogawa Y, Okabe M, Nishimune Y, Tanaka H. OAZ-t/OAZ3 is essential for rigid connection of sperm tails to heads in mouse. PLoS Genet 2009; 5(11): e1000712. 10.1371/journal.pgen.1000712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Devlin DJ, Agrawal Zaneveld S, Nozawa K, Han X, Moye AR, Liang Q, Harnish JM, Matzuk MM, Chen R. Knockout of mouse receptor accessory protein 6 leads to sperm function and morphology defects. Biol Reprod 2020; 102(6): 1234–1247. 10.1093/biolre/ioaa024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu F, Yan P, Zhang J, Cui Y, Zheng M, Cheng Y, Guo Y, Yang X, Guo X, Zhu H. Deficiency of TPPP2, a factor linked to oligoasthenozoospermia, causes subfertility in male mice. J Cell Mol Med 2019; 23(4): 2583–2594. 10.1111/jcmm.14149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castaneda JM, Miyata H, Archambeault DR, Satouh Y, Yu Z, Ikawa M, Matzuk MM. Mouse t-complex protein 11 is important for progressive motility in sperm. Biol Reprod 2020; 102(4): 852–862. 10.1093/biolre/ioz226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mi Y, Shi Z, Li J. Spata19 is critical for sperm mitochondrial function and male fertility. Mol Reprod Dev 2015; 82(11): 907–913. 10.1002/mrd.22536 [DOI] [PubMed] [Google Scholar]
- 55.Ma Q, Li Y, Luo M, Guo H, Lin S, Chen J, Du Y, Jiang Z, Gui Y. The expression characteristics of FAM71D and its association with sperm motility. Hum Reprod 2017; 32(11): 2178–2187. 10.1093/humrep/dex290 [DOI] [PubMed] [Google Scholar]
- 56.Nayernia K, Drabent B, Meinhardt A, Adham IM, Schwandt I, Muller C, Sancken U, Kleene KC, Engel W. Triple knockouts reveal gene interactions affecting fertility of male mice. Mol Reprod Dev 2005; 70(4): 406–416. 10.1002/mrd.20227 [DOI] [PubMed] [Google Scholar]
- 57.Miki K, Qu W, Goulding EH, Willis WD, Bunch DO, Strader LF, Perreault SD, Eddy EM, O’Brien DA. Glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme, is required for sperm motility and male fertility. Proc Natl Acad Sci U S A 2004; 101(47): 16501–16506. 10.1073/pnas.0407708101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Danshina PV, Geyer CB, Dai Q, Goulding EH, Willis WD, Kitto GB, McCarrey JR, Eddy EM, O’Brien DA. Phosphoglycerate kinase 2 (PGK2) is essential for sperm function and male fertility in mice. Biol Reprod 2010; 82(1): 136–145. 10.1095/biolreprod.109.079699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunleavy JEM, O’Bryan MK, Stanton PG, O’Donnell L. The cytoskeleton in spermatogenesis. Reproduction 2019; 157(2): R53–R72. 10.1530/REP-18-0457 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Sixty individually cropped PAS/H-stained seminiferous tubule cross-sections displaying the vacuolization abnormality widespread throughout Tssk3 KO mouse testes. This vacuolization is not present in controls. All panels at same magnification. Scale bar, 50 μm.
Supplementary Table 1.
Dataset of the quantitative profiling and the phosphoproteomics data.
Supplementary Table 2.
Differential phosphosites lists.


