Fig. 3.
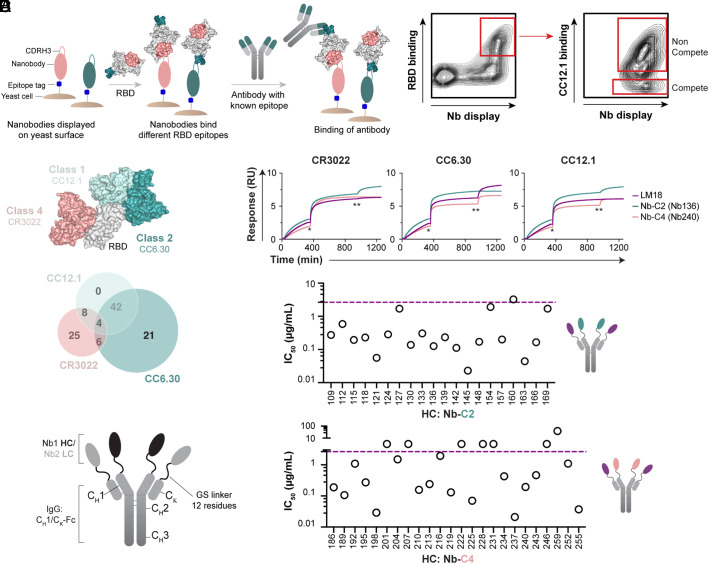
Design of bispecific antibodies and neutralization of Wuhan-1 SARS-CoV-2 PSV. (A) FACS-based epitope binning on yeast cell surface. RBD was first added to the induced yeast cells and binds to the nanobodies displayed on the surface. Then, the competitive antibody (CR3022, CC12.1, or CC6.30) was incubated and binds RBD only if the nanobody does not bind a similar epitope, or approaches RBD at an angle that is not blocked by the nanobody. (B) FACS plot showing the selection of competitive (C) and noncompetitive (NC) RBD-specific clones with CC12.1. (C) Nanobody binning based on C/NC profile and NGS analysis. Graphical representation of the three antibodies bound to RBD to show the location of their respective epitopes and Venn diagram with counts of competing nanobodies. (D) Confirmation of nanobody epitope by BLI-based assay. One representative for each of the class 2 (Nb-C2-136) and class 4 (Nb-C4-240) nanobodies is displayed. RBD was first added to the BLI sensor. Then, the competitive antibody (CR3022, CC12.1, or CC6.30) was then incubated (*) until signal saturation, followed by the tested nanobodies from our library (**). (E) Design of the bsNb4-Igs with one nanobody linked to CH1 and another nanobody linked to CL (kappa chain CK). (F) IC50 values (µg/mL) of bsNb4-Igs (Nb-C2 HC/LM18 LC, Top and Nb-C4 HC/ LM18 LC, Bottom) tested for neutralization of Wuhan-1 SARS-CoV-2 PSV. The IC50 value of LM18-Fc (2.65 µg/mL) is indicated by the purple dotted line.