Fig. 6.
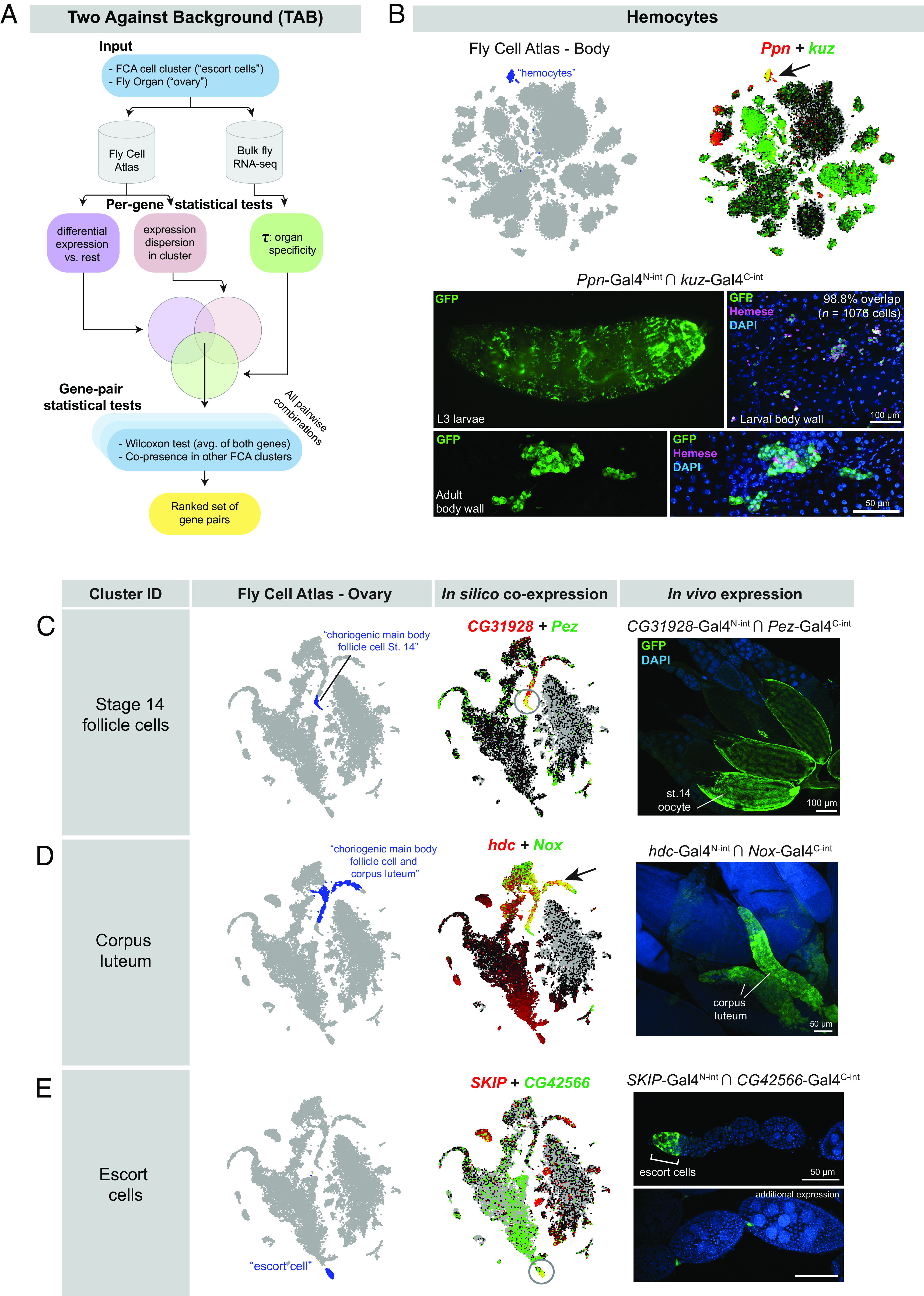
Characterization of split-intein Gal4 lines based on Two Against Background (TAB) algorithm predictions. (A) Schematic of the TAB algorithm to pick gene pairs that specifically mark scRNAseq clusters. (B) Hemocyte-specific split-intein Gal4 line based on TAB gene pairs. Top Left: Fly Cell Atlas 10× “Body” atlas, with hemocyte cluster indicated in blue. Top Right: In silico prediction of Ppn and kuz coexpression in the hemocyte cluster, with coexpression shown in yellow. Bottom: In vivo expression of the Ppn ∩ kuz split-intein Gal4 driver in larval and adult hemocytes. Hemocytes are costained with the H2 antibody against Hemese, a larval pan-hemocyte marker. (C–E) In silico predictions of coexpression in three different clusters from the FCA “Ovary” dataset and in vivo coexpression in the indicated cell types of the ovary. scRNAseq data are screenshots from the FCA data viewer for the “10×, stringent” datasets.
