Abstract
The amino acid sequence of human cytoplasmic cysteinyl-tRNA synthetase (CRS) was examined by analyzing sequences of genomic and expressed sequence tag fragments. From theses analyses, a few interesting possibilities were suggested for the structure of human CRS. First, different isoforms of CRS may result from alternative splicing. Second, the largest one would comprise 831 amino acids. Third, a new exon was identified encoding an 83 amino acid domain that is homologous to parts of elongation factor-1 subunits as well as other proteins involved in protein synthesis. Northern blot analysis showed three different mRNAs for CRS (of ∼3.0, 2.7 and 2.0 kb) from human testis while only the 2.7 kb mRNA was commonly detected in other tissues. Expression of the exon 2-containing transcript in testis was confirmed by RT–PCR and northern blotting. CRS containing the exon 2-encoded peptide retained catalytic activity comparable to that lacking this peptide. This peptide was responsible for the specific interaction of CRS with elongation factor-1γ.
INTRODUCTION
Aminoacyl-tRNA synthetases (ARSs) decode genetic information into amino acids with high fidelity. Since their catalytic activities are essential for protein synthesis and cell viability, they are classified as housekeeping enzymes. However, they are also involved in various biological processes (1). The functional diversity of these enzymes appears to be related to their structural characteristics. In particular, mammalian ARSs show interesting features in their structure and expression. They have non-catalytic extensions and form macromolecular complexes with each other (2–4) or with other cellular molecules (5–8). Expression of mammalian tryptophanyl-tRNA synthetase is induced by interferon-γ (9) and its isoforms are generated by alternative splicing (10). Generation of different transcripts by alternative splicing was also suggested for human isoleucyl-tRNA synthetase (11). Signal-dependent induction and the presence of isoforms also support the novel biological functions of ARSs.
In the present work, we examined cDNA, genomic DNA and expressed sequence tag (EST) fragments for human cysteinyl-tRNA synthetase (CRS) and found the presence of different isoforms. The first reported amino acid sequence for human CRS (P49589) is composed of 638 amino acids (12). Later, the cDNA sequence for human CRS was deposited in GenBank by the same authors but it lacks the 5′-end encoding the N-terminal 43 amino acids (L06845). Human CRS contains the class I signature motifs HIGH and KMSKS in the catalytic domain (13–15) and shows moderate homology to CRSs of other species. However, the deposited sequence is devoid of the C-terminal extension that is found in other eukaryotic CRSs (Fig. 1, Homo sapiens 1). The cDNA sequence for human CRS was then revised to comprise 750 amino acids (85.7 kDa) with the C-terminal extension (16; Fig. 1, H.sapiens 2). Although this new sequence was more consistent with yeast CRS (17) in size and sequence, it still appeared to contain sequence errors to be fixed (data not shown). In addition, CRSs of 86 and 92 kDa were isolated from rabbit liver (16). Although the revised human CRS fits with the smaller rabbit CRS, the source of the larger one has not been explained. Finally, different CRS isoforms were also identified from rat (18), despite the fact that only one CRS gene was identified (19). For these reasons, the structural gene for human CRS needs to be re-examined.
Figure 1.
Sequence alignment of CRSs from different species. Schematic alignment of representative CRSs. Eukaryotic CRSs, compared to prokaryotic (Escherichia coli) and archeaic (Aeropyrum pernix) CRSs, have two characteristics, a large insertion of ∼100 residues within a Rossmann fold and an extension at the C-terminus. The reported sequence of human CRS (P49589, H.sapiens 1) lacks the C-terminal extension (12) but was revised to have the C-terminal extension (H.sapiens 2) (16). These two sequences are to be corrected.
First, we analyzed the exon–intron arrangement of the genomic sequence of human CRS and also collected EST fragments to re-assemble the cDNA for human CRS. From these analyses, different combinations of exons and a new potential exon were implied. This prediction was confirmed experimentally by northern blotting and RT–PCR. The newly identified exon encodes the peptide homologous to elongation factor (EF) subunits 1β and 1γ. The functional significance of this inserted domain was further investigated.
MATERIALS AND METHODS
Analysis of genomic and EST sequences of human CRS
The cDNA (L06845) and genomic sequences (AC001228) of human CRS were obtained from GenBank. The reported CRS cDNA lacks 129 nt encoding the N-terminal 43 amino acids. The missing 5′-end of the cDNA sequence was found by searching a human EST database using BLASTN (20). The exon–intron arrangement was determined by comparing the cDNA and genomic sequences and was also predicted using the GENSCAN (21) and FGENE programs (22). A total of 116 EST sequences of human CRS were collected from the database and assembled to reconstitute the full-length cDNA. Comparison of nucleotide sequences was performed using the BLAST 2 Sequences service (23) provided by the National Center for Biotechnology Information of the National Institutes of Health. Multiple sequence alignments of proteins were performed using the program CLUSTALX (24).
Isolation of the 5′-end of cDNA for human CRS
The PCR primers C5F (5′-CCGTCGACATGGCAGATTCCTCCGGGCAGCAG) and C5R (5′-GTTCACAATTTCTGGCACATACTC) were synthesized based on the known 5′ cDNA sequence of human CRS and used to re-isolate cDNA encoding human CRS from a human testis cDNA library (Marathon cDNA; Clontech, CA, USA). The PCR reaction was carried out using High Fidelity Taq polymerase following the manufacturer’s protocol (Boehringer Mannheim, Mannheim, Germany). The PCR products were phosphorylated, filled-in and ligated to SmaI cut pTZ19R (Amersham-Pharmacia Biotech, Uppsala, Sweden).
Poly(A) RNA preparations of human testis, thymus and intestine (Clontech) were used for RT–PCR using specific primer pairs to isolate various fragments derived from the cDNA of human CRS. Each poly(A) RNA was converted to cDNA by reverse transcription. The reaction was carried out at 42°C for 1 h using AMV reverse transcriptase (Boehringer Mannheim) following the manufacturer’s instructions in the presence of poly(A) RNA (0.2 µg), RNase inhibitor (20 U) and the pairs of specific primers. The reaction was stopped by heat inactivation of reverse transcriptase at 75°C for 15 min. The cDNA obtained was amplified by PCR using High Fidelity Taq polymerase as described above and analyzed by agarose gel electrophoresis.
Construction of human CRS cDNAs
Recombinant plasmid pKS583 (a derivative of pET19a) contains the cDNA of human CRS as previously reported (12). To prepare the new cDNA encoding exon 2+ CRS, the cDNA fragment containing exon 2 was generated by PCR from the human testis cDNA library, digested with SalI and DraIII and ligated into the same sites of pKS583. The resulting plasmid was designated pKS583-1.
Northern blotting
Expression of CRS was determined by northern blotting with a probe specific to exon 2 or the common region of human CRS. The probes were prepared by PCR using the specific primers and purified by gel elution. The purified probes (150–200 ng) were labeled by Klenow fragment at 30°C for 4 h in the presence of [α-32P]dATP and 2 mM each dGTP, dCTP and dTTP. The labeled probes were purified using a microspin column (Micro Bio-spin 25P-6 columns; Bio-Rad, CA, USA). These probes were hybridized to RNAs on a multiple tissue Northern Blot II (MTN‘; Clontech) following the manufacturer’s instructions. The hybridized probes were detected by autoradiography.
Yeast two-hybrid assay
Interactions of CRS with EF-1 subunits and p18 were tested in the yeast two-hybrid system (25). Human CRS (exon 2+ or exon 2–) cDNA was cut out from pKS583 or pKS583-1 by digestion with SalI and NotI and inserted into pCMXGal4N to generate appropriate restriction sites. To express LexA–CRS, the CRS gene was re-isolated from pCMXGal4N-CRS with EcoRI (partial digestion) and NotI and ligated into pEG202 digested with the same enzymes. The cDNAs encoding the subunits of human EF complex I were obtained by PCR with the corresponding primers. A human lymphocyte cDNA library (Clontech) was used as template for cloning cDNAs for EF-1α1 and EF-1α2, IMAGE clones 1736996 and 50603 for EF-1β and EF-1δ, respectively, and a human fetal brain cDNA library (Clontech) for EF-1γ. The cDNA of p18 was isolated from pM421 (H.Motegi, Cancer Institute, Japan) by PCR using specific primers. These cDNAs were subcloned into the EcoRI and XhoI sites of pJG4-5 and the proteins expressed as B42 fusion proteins. In the two hybrid proteins, LexA and B42 function as the specific DNA-binding and transcription activation domains, respectively and a positive interaction between the attached proteins X and Y was determined by induction of β-galactosidase as described previously (26).
Aminoacylation activity
The cDNAs encoding CRS with and without exon 2 were cloned into pcDNA3 (Myc-Flag tagged) using EcoRI (partial cut) and NotI. The recombinant plasmids and vector alone were transfected into 293 cells grown on 150 mm plates using GenePORTER (Gene Therapy Systems, CA, USA). The cells were harvested 48 h after transfection and suspended in 2 ml of 20 mM Tris–HCl, pH 7.5, 20 mM KCl, 10 mM MgCl2, 5 mM dithiothreitol (DTT), leupeptin, pepstatin A, aprotinin (5 µg/ml each) and 300 µM phenylmethylsulfonyl fluoride. The cells were then homogenized and centrifuged at 5000 g for 40 min and supernatants were used for aminoacylation. The reaction was initiated by adding the supernatants (2 µg total protein) to 50 µl of 20 mM Tris–HCl, pH 7.5, 2 mM ATP, 20 mM KCl, 10 mM MgCl2, 5 mM DTT, 35 µg calf liver tRNA (Boehringer Mannheim), 10 µCi [35S]cysteine (>600 Ci/mmol; NEN, MA, USA). The reactions were sampled at 1 min intervals and stopped with 10% trichloroacetic acid (TCA) on 2.3 cm glassfiber filter disks (Whatman, Kent, UK). After washing the filters with 5% cold TCA and 95% ethanol, aminoacylated cysteine was quantified by a liquid scintillation counter (1409 DSA; Wallac, Turku, Finland).
In vitro pull-down assay
EF-1γ fused to GST or GST alone were expressed in Escherichia coli BL21 (DE3). The E.coli protein extract containing GST-EF-1γ or GST was first mixed with glutathione–Sepharose in phosphate-buffered saline at 25°C for 15 min. The exon 2+ CRS was synthesized by in vitro translation in the presence of [35S]methionine using pcDNA3-exon 2+ CRS. The in vitro translated exon 2+ CRS was added to GST proteins with the E.coli BL21 (DE3) extract and incubated at 4°C for 4 h with rotation in binding buffer of 40 mM HEPES, pH 7.6, 20 mM NaCl, 2 mM EDTA, 20% glycerol, 1 mM DTT, 300 µM phenylmethylsulfonyl fluoride and protease inhibitor cocktail (pepstatin A, leupeptin, antipain and chymostatin, 5 µg/ml each). The reaction mixture was then washed three times with binding buffer containing 0.1% Triton X-100 and the proteins bound to the Sepharose beads were eluted with SDS sample buffer for electrophoresis. The eluted proteins were separated by SDS gel electrophoresis and the presence of the exon 2+ CRS was determined by autoradiography.
RESULTS
Genomic and EST sequence analyses of human CRS
Sequence alignments of 27 known CRSs revealed that eukaryotic CRSs have large insertions within the Rossmann fold and C-terminal extensions compared to prokaryotic and archeaic CRSs (Fig. 1). The C-terminal extensions from Saccharomyces cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans and Drosophila melanogaster exhibit high sequence identities and similarities (>31 and 54%, respectively). The human CRS sequence in GenBank (P49589) lacks a similar C-terminal extension (Fig. 1, H.sapiens 1) but was later revised to contain the longer C-terminal peptide (16; Fig. 1, H.sapiens 2).
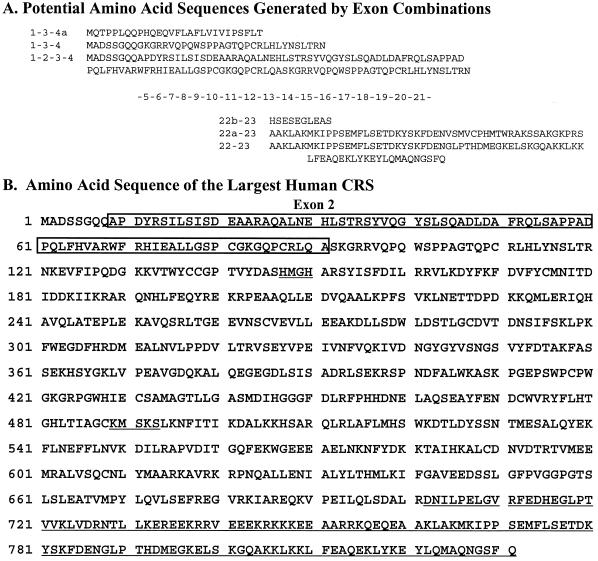
The structural gene for human CRS is located on a 244 kb contig of human chromosome 11p15.5 (AC001228). A comparison between the cDNA and genomic sequences revealed 22 exons and several discrepancies, such as deletions, insertions and mutations (data not shown). To further study the exon–intron arrangement, two exon prediction programs were employed. The programs GENSCAN (21) and FGENE (22) predicted 30 and 28 exons, respectively, most of which were consistent with those present in the cDNA sequence (data not shown). In addition, a total of 116 EST fragments of human CRS were collected from the database and assembled. From these analyses, a few interesting points were suggested. First, alternative splicing may occur in the expression of human CRS at the 5′- and 3′-ends of the transcript (Fig. 2A). Second, a new exon (designated exon 2) was predicted by both of the exon analysis programs. This exon consists of 249 nt encoding 83 amino acids in-frame with the known amino acid sequence of human CRS (Fig. 2A and B). Third, the largest CRS that can be produced by alternative splicing would comprise 831 amino acids, which is larger than the size of the previously reported human CRS (Fig. 2B). Here, we have focused on exon 2 and investigated the expression and functional significance of this exon.
Figure 2.
Potential CRS isoforms and deduced amino acid sequence of the largest human CRS. (A) Amino acid sequences are shown based on potential combinations of exons. Analyses of genomic DNA, cDNA and EST sequences reveal 23 exons; one newly predicted exon (exon 2) and two alternatively spliced exons. Exon 4 has two splicing start points (4 and 4a) and exon 22 has two splicing start points and two end points (22, 22a, 22b and 22c). Thus, the N-terminus has four possible combinations of exon 2, exons 2–, 2+, 4 and 4a, and the C-terminus has four possible forms of exon 22. Translation start positions are different for combinations of 1-3-4 and 1-3-4a. Exon combinations 1-2-3-4a and 22c-23 are not shown because they would give the same results as sequences 1-3-4a and 22b-23, respectively. (B) The deduced amino acid sequence of the largest human CRS. A transcript containing exons 2, 4 and 22 would yield an 831 amino acid polypeptide. The residues from exon 2 are boxed and the C-terminal sequence different from the corresponding region of CRS in GenBank is underlined. The two class I signature sequences (HMGH and KMSKS) in the core domain are also underlined.
Identification of exon 2 in the transcript for human CRS
The 5′-end fragment of cDNA for human CRS was isolated by PCR using a human testis cDNA library as template with specific primers. Based on the known cDNA for CRS, PCR with the pair of primers C5F and C5R is supposed to generate a 758 bp DNA fragment of the 5′-end. Two DNA fragments of ∼1.0 and 0.7 kb were obtained by PCR (Fig. 3). Sequencing of the two fragments showed that the larger one contained an additional sequence of 249 bp corresponding to exon 2. This result confirmed the expression of exon 2 that was predicted by sequence analyses (Fig. 2).
Figure 3.
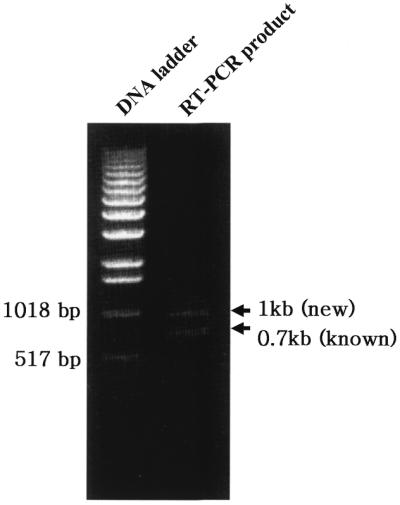
Isolation of 5′-end cDNA for human CRS. A human testis cDNA library was used as template for PCR to amplify the 5′-end of cDNA encoding CRS. The primers specific to the 5′-end of CRS cDNA were used for the experiments. Two different PCR products of ∼1.0 and 0.7 kb were generated.
Expression of CRS isoforms
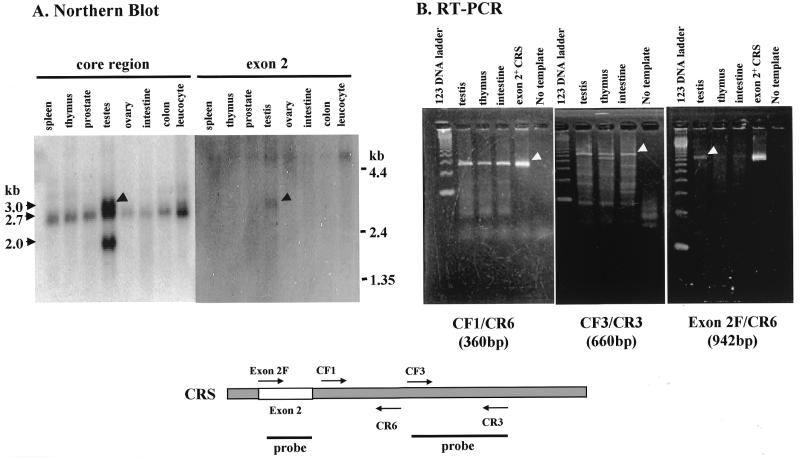
To determine whether CRS containing the exon 2 peptide is expressed in specific tissues, northern blotting and RT–PCR were carried out. The mRNAs isolated from various human tissues were first hybridized with the radioactive probes prepared from the core domain of CRS. While mRNA of ∼2.7 kb was detected from most of the tissues, the larger and smaller mRNAs of ∼3.0 and 2.0 kb were additionally detected in testis (Fig. 4A, left). Then, the same mRNA membrane was re-hybridized with the exon 2 probe. The probe hybridized only with the mRNA of 3.0 kb, indicating that this mRNA contains exon 2. The smallest mRNA appears to be one of 2255 bp encoding the shortest CRS polypeptide of 676 amino acids (Fig. 2A). Northern blotting was also conducted with the probe for human methionyl-tRNA synthetase but multiple bands were not detected in any of the tissues (data not shown).
Figure 4.
Expression of exon 2+ CRS in testis. (A) The mRNAs isolated from various human tissues were hybridized with the radioactively labeled probes for the core region or for exon 2 of CRS (indicated below). (B) The mRNAs isolated from human testis, thymus and intestine were subjected to RT–PCR using the indicated pairs of primers specific to the different regions of mRNA for human CRS (indicated below).
To confirm this pattern of expression, we carried out RT–PCR of mRNAs isolated from human testis, thymus and intestine with pairs of primers specific to the different regions of human CRS. The pairs CF1/CR6 and CF3/CR3 should generate 360 and 660 bp DNA fragments from the core region of CRS. DNA fragments of the expected sizes were produced from all of the three mRNAs. Then, the primer specific for the sequence of exon 2 was combined with CR6 for RT–PCR. The PCR product was generated from mRNA of testis but not from those of thymus and intestine (Fig. 4B). This result also confirmed the specific and abundant expression of exon 2+ CRS in testis.
Aminoacylation activity of CRS isoforms
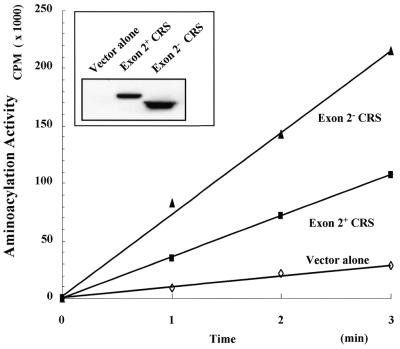
To determine whether the two CRS isoforms were both catalytically active, they were expressed in mammalian cells as Myc-tagged protein and the enzyme activity determined. Expression of the two proteins was confirmed by immunoblotting with anti-Myc antibody (Fig. 5, inset). Exon 2+ CRS showed catalytic activity at ∼50% of the level of exon 2– CRS (Fig. 5). This result suggests that the inserted domain expressed from exon 2 is not essential for the enzyme activity of CRS.
Figure 5.
Aminoacylation activity of exon 2+ and exon 2– CRS. 293 cells were transfected with pcDNA3 alone or containing the cDNAs coding for exon 2+ or exon 2– CRS. The cells were harvested 48 h after transfection and lysed as described in Materials and Methods. Expression of two CRS isoforms was determined by immunoblotting with anti-Myc antibody (inset) and the lysates were used for aminoacylation assay. The aminoacylation activity of the two CRS isoforms was calibrated by expression level of each protein.
Association of exon 2+ CRS with EF-1γ
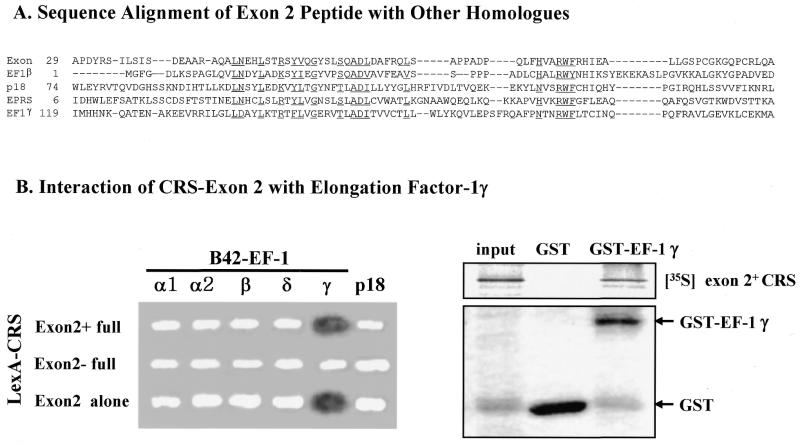
The amino acid sequence encoded by exon 2 showed homology with parts of human EF-1 subunits β and γ and other proteins, including p18 (27) and human glutamyl-prolyl-tRNA synthetase (EPRS), which are components of the multi-ARS complex (2–4; Fig. 6A). Subunits β and δ of EF-1 form a complex with subunit γ and stimulate EF-1α activity (28). In addition, it was previously reported that some mammalian ARSs associate with the EF-1 complex to facilitate delivery of aminoacylated tRNA to the ribosome for protein synthesis (5–7). We thus tested whether the peptide encoded by exon 2 is responsible for interaction of CRS with the EF-1 complex in a yeast two-hybrid system (25). Exon 2+ CRS, exon 2– CRS and exon 2 alone were fused to the DNA-binding domain LexA and the subunits of EF-1 and p18 were fused to the transcriptional activator domain B42. A positive interaction was determined by induction of the reporter gene β-galactosidase. A strong and specific interaction was detected for the pairs of exon 2 alone or exon 2+ CRS with EF-1γ (Fig. 6B, left), indicating that the peptide encoded by exon 2 is responsible for the association of CRS with EF-1. The interaction of exon 2 peptide and EF-1γ was also confirmed by in vitro pull-down assay. GST or GST–EF-1γ was mixed in vitro with translated 35S-labeled exon 2+ CRS. The exon 2+ CRS was co-eluted with GST–EF-1γ but not with GST alone (Fig. 6B, right).
Figure 6.
Sequence alignment of the peptide encoded by exon 2 of CRS and its interaction with EF-1γ. (A) The peptide of CRS exon 2 shows sequence homology with those of human elongation factor 1β (43% over 53 residues), human EPRS (glutamyl-prolyl-tRNA synthetase) (43% over 55 residues), p18 (30% over 55 residues) and human EF-1γ (30% over 55 residues). Multiple sequence alignment of the above five proteins was prepared by CLUSTALX and the conserved residues are underlined. (B) (Left) Interactions of the peptide encoded by CRS exon 2 with the subunits of EF-1 and p18 (27) were tested by yeast two-hybrid assay. Exon 2+ CRS, exon 2– CRS and exon 2 alone were expressed as hybrid proteins with LexA and the EF-1 subunits and p18 were fused to B42. A positive interaction was determined as a blue color (shown as dark color in the print) on X-gal-containing medium. (Right) The interaction of exon 2+ CRS with EF-1γ was tested by in vitro pull-down assay. The in vitro translated 35S-labeled exon 2+ CRS was mixed with GST–EF-1γ or GST alone and bound to glutathione–Sepharose. The proteins eluted from the Sepharose beads were separated by SDS gel electrophoresis and the radioactive exon 2+ CRS was detected by autoradiography.
DISCUSSION
Although ARSs play a housekeeping role as catalysts, control of their expression is rather complicated in some cases. Mammalian tryptophanyl-tRNA synthetase is induced by interferon (9) and four different ARSs are expressed at high level in the salivary gland of Drosophila (29). The expression of ARSs is also regulated by alternative splicing. Two and four different mRNA isoforms have been found for human isoleucyl-tRNA synthetase (11) and tryptophanyl-tRNA synthetase (10), respectively. Interestingly, sequences homologous to an interferon-stimulated response element have commonly been found in the regulatory regions of these genes (11,30), although the relationship between this element and alternative splicing is not understood. Here, we have identified a new exon for human CRS and suggested alternative splicing to express different isoforms of this enzyme.
The genomic sequences of five different ARSs (cysteinyl-, valyl-, histidyl- and glycyl-tRNA synthetase and phenylalanyl-tRNA synthetase α subunit) are currently available. We have analyzed their genomic sequences to search for possible alternative splicing. Among these genes, different forms of transcript were suggested only for CRS (data not shown), implying that alternative splicing is not universal in the expression of ARSs (data not shown). Different amino acid sequences were predicted for both ends of CRS (Fig. 2A). In fact, an mRNA smaller than the previously reported CRS was also isolated from the 3′-end by RT–PCR (data not shown). The sequence of this transcript matched the exon combination 22b and 23 (Fig. 2A), verifying the prediction of exon combination. A total of three combinations of exons are possible in the N- and C-terminal ends (Fig. 2A), but we do not know whether all of these combinations really take place in the cell. The largest CRS containing the peptide of exon 2 is composed of 831 amino acids (expected mol. wt 94.6 kDa).
The peptide encoded by exon 2 of CRS is not essential for catalysis because CRS was active regardless of its presence (Fig. 5). Thus, the biological significance of this peptide is not yet clear. Interestingly, the exon 2 peptide contains a domain homologous to subunits β and γ of EF-1 (Fig. 6A) and interacts with EF-1γ (Fig. 6B). It was previously shown that mammalian valyl- and aspartyl-tRNA synthetases interact with the EF-1 complex (5–7). The interactions of ARSs with EF stimulate aminoacylation by facilitating vectoral transfer of aminoacylated tRNAs (6,31,32). By analogy, the role of the exon 2 peptide may facilitate delivery of cognate tRNA from CRS to EF. Thus, exon 2+ CRS may show much higher cellular activity than exon 2– CRS, although the two forms of CRS showed comparable in vitro activity (Fig. 5). The enhanced activity of CRS may be required to supply cysteinylated tRNA for the synthesis of cysteine-rich testis proteins such as metallothionein (33). However, it is also possible that exon 2+ CRS may have a novel function not directly related to its catalytic activity.
The domain homologous to the subunits of EF-1 was also found in p18 and the N-terminal region of glutamyl-prolyl-tRNA synthetase (Fig. 6A). p18 is one of the auxiliary factors associated with the multi-ARS complex (27). This protein also interacts with EF-1 (data not shown). Thus, mammalian ARSs appear to be structurally and functionally linked to the EF complex via their non-catalytic domains (exon 2 peptide of CRS; the N-terminal extensions of valyl- and aspartyl-tRNA synthetases) as well as via adaptors such as p18. The association of ARSs and the EF complex may be necessary to constitute the higher order complex of the protein synthesis machinery or to control the cellular localization of these proteins.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the National Creative Research Initiatives of the Ministry of Science and Technology of Korea to S.K. and from Hallym Academy of Science, Hallym University.
REFERENCES
- 1.Martinis S.A., Plateau,P., Cavarelli,J. and Florentz,C. (1999) EMBO J., 18, 4591–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirande M. (1991) Prog. Nucleic Acid Res. Mol. Biol., 40, 95–142. [DOI] [PubMed] [Google Scholar]
- 3.Kisselev L.L. and Wolfson,A.D. (1994) Prog. Nucleic Acid Res. Mol. Biol., 48, 83–142. [DOI] [PubMed] [Google Scholar]
- 4.Yang D.C.H. (1996) Curr. Top. Cell. Regul., 34, 101–136. [DOI] [PubMed] [Google Scholar]
- 5.Brandsma M., Kerjan,P., Dijk,J., Janssen,G.M.C. and Moller,W. (1995) Eur. J. Biochem., 233, 277–282. [DOI] [PubMed] [Google Scholar]
- 6.Negrutskii B.S., Shalak,V.F., Kerjan,P., El’skaya,A.V. and Mirande,M. (1999) J. Biol. Chem., 274, 4545–4550. [DOI] [PubMed] [Google Scholar]
- 7.Reed V.S. and Yang,D.C.H. (1994) J. Biol. Chem., 269, 32937–32941. [PubMed] [Google Scholar]
- 8.Park S.G., Jung,K.H., Lee,J.S., Jo,Y.J., Motegi,H., Kim,S. and Shiba,K. (1999) J. Biol. Chem., 274, 16673–16676. [DOI] [PubMed] [Google Scholar]
- 9.Kisselev L., Frolova,L. and Haenni,A.L. (1993) Trends Biochem. Sci., 18, 263–267. [DOI] [PubMed] [Google Scholar]
- 10.Pajot B., Sarger,C., Bonnet,J. and Garret,M. (1994) J. Mol. Biol., 242, 599–603. [DOI] [PubMed] [Google Scholar]
- 11.Nichols R.C., Raben,N., Boerkoel,C. and Plotz,P.H. (1995) Gene, 155, 299–304. [DOI] [PubMed] [Google Scholar]
- 12.Cruzen M.E. and Arfin,S.M. (1994) DNA Seq., 4, 243–248. [DOI] [PubMed] [Google Scholar]
- 13.Webster T.A., Tsai,H., Kula,M., Mackie,G.A. and Schimmel,P. (1984) Science, 226, 1315–1317. [DOI] [PubMed] [Google Scholar]
- 14.Ludmerer S.W. and Schimmel,P. (1987) J. Biol. Chem., 262, 10801–10806. [PubMed] [Google Scholar]
- 15.Hountondji C., Dessen,P. and Blanquet,S. (1986) Biochimie, 68, 1071–1078. [DOI] [PubMed] [Google Scholar]
- 16.Motorin Y. and Waller,J.P. (1998) Biochimie, 80, 579–590. [DOI] [PubMed] [Google Scholar]
- 17.Motorin Y., Le Caer,J.P. and Waller,J.P. (1997) Biochimie, 79, 731–740. [DOI] [PubMed] [Google Scholar]
- 18.Pan F., Lee,H.H., Pai,S.H., Yu,T.C., Guoo,J.Y. and Duh,G.M. (1976) Biochim. Biophys. Acta, 452, 271–283. [DOI] [PubMed] [Google Scholar]
- 19.Cruzen M.E., Bengtsson,U., McMahon,J., Wasmuth,J.J. and Arfin,S.M. (1993) Genomics, 15, 692–693. [DOI] [PubMed] [Google Scholar]
- 20.Altschul S.F., Gish,W., Miller,W., Myers,E. and Lipman,D.J. (1990) J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 21.Burge C. and Karlin,S. (1997) J. Mol. Biol., 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 22.Solovyev V.V., Salamov,A.A. and Lawrence,C.B. (1994) Nucleic Acids Res., 22, 5156–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatusova T.A. and Madden,T.L. (1999) FEMS Microbiol. Lett., 174, 247–250. [DOI] [PubMed] [Google Scholar]
- 24.Tompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gyuris J., Golemis,E., Chertkov,H. and Brent,R. (1993) Cell, 75, 791–803. [DOI] [PubMed] [Google Scholar]
- 26.Rho S.B., Kim,M.J., Lee,J.S., Seol,W., Montegi,H., Kim,S. and Shiba,K. (1999) Proc. Natl Acad. Sci. USA, 96, 4488–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quevillon S. and Mirande,M. (1996) FEBS Lett., 395, 63–67. [DOI] [PubMed] [Google Scholar]
- 28.Sheu G.-T. and Traugh,J.A. (1997) J. Biol. Chem., 272, 33290–33297. [DOI] [PubMed] [Google Scholar]
- 29.Seshaiah P. and Andrew,D.J. (1999) Mol. Biol. Cell, 10, 1595–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleckner J., Rasmussen,H.H. and Justesen,J. (1991) Proc. Natl Acad. Sci. USA, 88, 11520–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Negrutskii B.S., Budkevich,T.V., Shalak,V.F., Turkovskaya,G.V. and El’Skaya,A.V. (1996) FEBS Lett., 382, 18–20. [DOI] [PubMed] [Google Scholar]
- 32.Reed V.S., Wasteny,M.E. and Yang,D.C.H. (1994) J. Biol. Chem., 269, 32932–32936. [PubMed] [Google Scholar]
- 33.Hunziker P.E. and Kagi,J.H. (1985) Biochem. J., 15, 375–382. [DOI] [PMC free article] [PubMed] [Google Scholar]