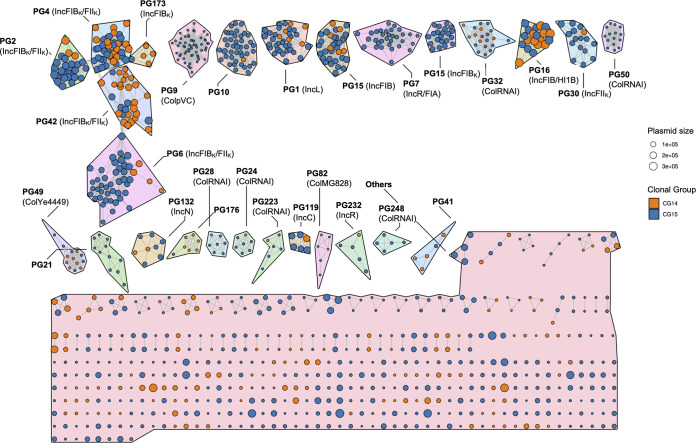
FIG 3.
K-nearest neighbor network (K-NNN) of K. pneumoniae CG14 and CG15 plasmids. The network was built using the PATO k-nnn function. Each node represents one plasmid and is colored according to the CG. A group of similar plasmids (>5 plasmids) is represented by a plasmid group (PG), plasmid clusters being defined using the Louvain algorithm over the network structure. Each PG is arbitrarily designated with a number, and replicon content defined by MOB-typer is shown in parentheses (34). Differences in color shading between PGs is arbitrary. Each plasmid is connected with the 10 best hits if their Jaccard similarity is at least 0.5 and the difference in size is less than 50%.