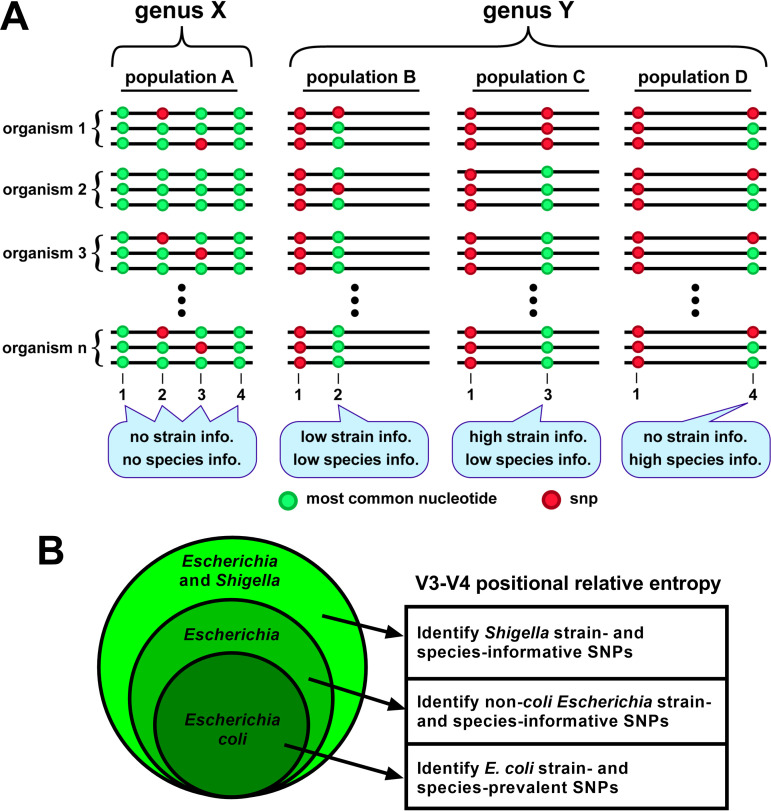
FIG 1.
Illustration of 16S rRNA V3-V4 positional relative entropy. Relative entropy analysis can be used to identify strain- or species-specific residues among variable region sequences in a population. (A) Schematic depictions of variable regions of a multi-copy gene found in 2 hypothetical genera, X, and Y. In a sequenced cohort of genus X organisms (population A), invariant residues at positions 1 and 4 provide no sub-genus information, and SNPs observed at positions 2 and 3 are not associated with a particular species or strain, so their identities provide no information at those taxonomic levels. In genus Y, the SNP at position 1 is a strong genus indicator (relative to that of genus X). In population B, occasional SNPs at position 2 provide no information because they are observed in single alleles in strains nonspecific to a species. In population C, an SNP at position 3 is a strong strain indicator because it is present in all alleles in the strain’s genome. In population D, an occasional SNP at position 4 indicates the presence of that species but provides no strain information. (B) A Venn diagram illustrating the process of using positional relative entropy (DKL) to identify informative V3-V4 residues, starting from (i) those prevalent in a strain or multiple strains relative to the total E. coli population, then (ii) those informative of non-coli Escherichia strains and species relative to the total Escherichia population, and lastly (iii) those informative of Shigella strains and species relative to the total Escherichia and Shigella population.