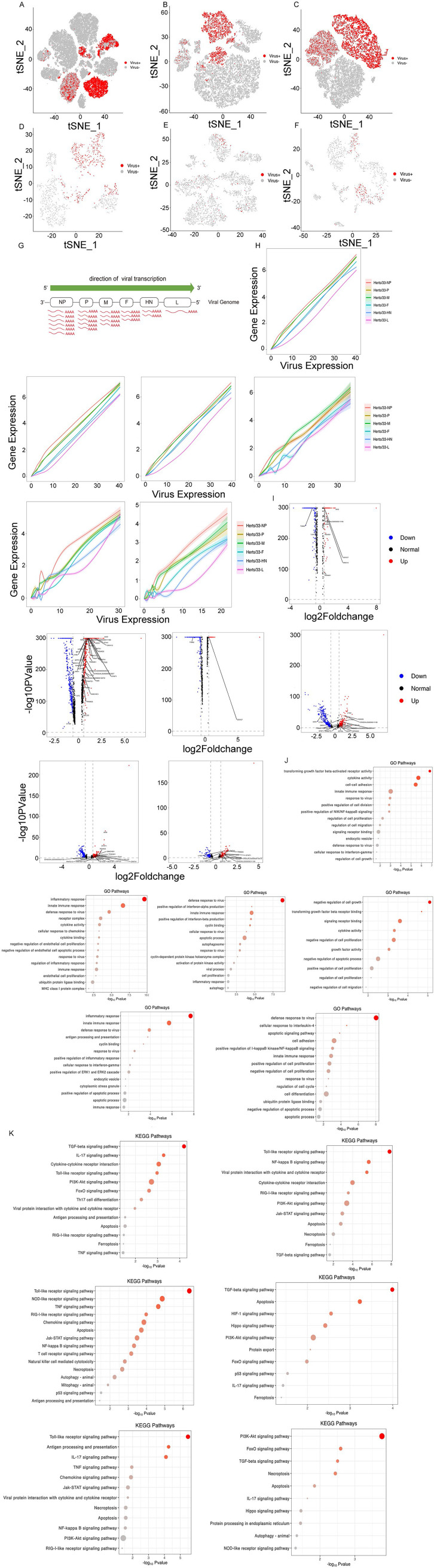
FIG 7.
Cell types with highly virulent NDV Herts/33 strain RNA detected. (A) A total of 56,995 cells with Herts/33 RNA detected (UMI counts of >0) from the total cells both in vitro and in vivo. (B to F) Cells with Herts/33 RNA detected (UMI counts of >0) from fibroblast, myeloid, endothelial, epithelial, and T cells. (G) Schematic of NDV transcription. The viral RNA-directed RNA polymerase transcribes each gene sequentially but occasionally releases the genomic RNA template, ending transcription. Thus, the transcription frequency decreases from NP to L. (H) Proportion of each NDV gene versus the viral load from total, fibroblast, myeloid, endothelial, epithelial, and T cells. The x axis indicates the sum of the expression of six viral genes in each cell, and the y axis indicates the expression of each viral gene in each cell. (I) Volcano plots showing DEGs of virus-positive versus virus-negative cells from total, fibroblast, myeloid, endothelial, epithelial, and T cells. ISGs are shown in the plot (P < 0.05). Red indicate up regulation and blue indicated down regulation. (J and K) GO and KEGG pathway analyses of IFN signaling and the response to virus signaling, comparing virus-positive to virus-negative cells from total, fibroblast, myeloid, endothelial, epithelial, and T cells.