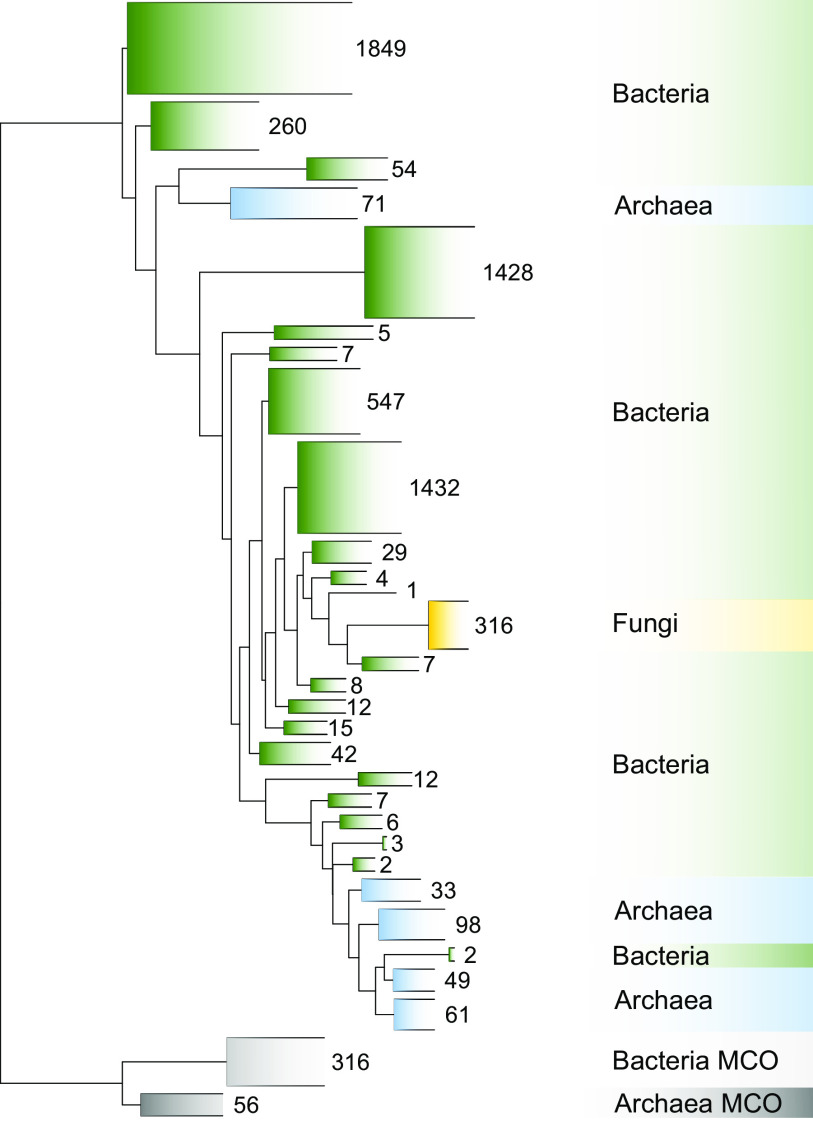
FIG 2.
Reference phylogeny of prokaryotic and fungal nirK, created from 6,732 full-length genomic nirK gene sequences. The phylogeny was determined based on a maximum likelihood analysis of the amino acid sequences of nirK, using the LG+G substitution model. The phylogeny was subsequently used as the reference tree for the phylogenetic placement of nirK gene fragments that were retrieved from 1,980 metagenomes. Clades were collapsed, and the number of species per collapsed clade is indicated. The outgroups consist of multicopper oxidase (MCO) sequences originating from cyanobacterial and thaumarcheotal genomes.