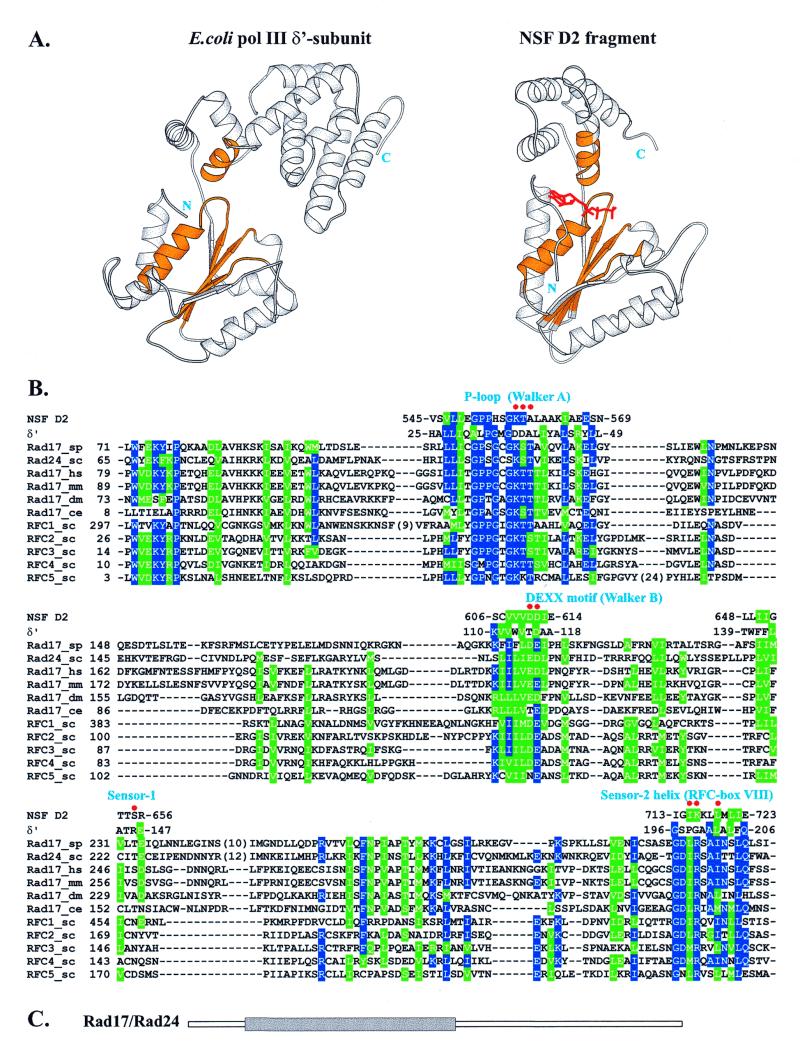
Figure 3.
(A) Comparison of 3D structures of the δ′ subunit of E.coli polymerase III and the NSF D2 fragment. Equivalent structural motifs involved in forming the nucleotide-binding site that could also be unambiguously aligned with the Rad17 and RFC families are colored gold. The ATP analog bound to NSF D2 is shown in red. (B) Alignment of the Rad17 family and all five yeast RFC subunits with the structural fragments of both δ′ and NSF D2, colored gold in (A), subsequently used to construct the model for Rad17Sp given in Figure 4. Residues conserved in more than half of the sequences are colored blue (identical) and green (similar). Red dots indicate Rad17Sp nucleotide-binding site residues shown with their side chains in Figure 4. Numbers indicate positions of residues in sequences. (C) Location of the Rad17 (Rad24Sc) region (gray rectangle) aligned in (B), found to be common to the AAA+ class (39).