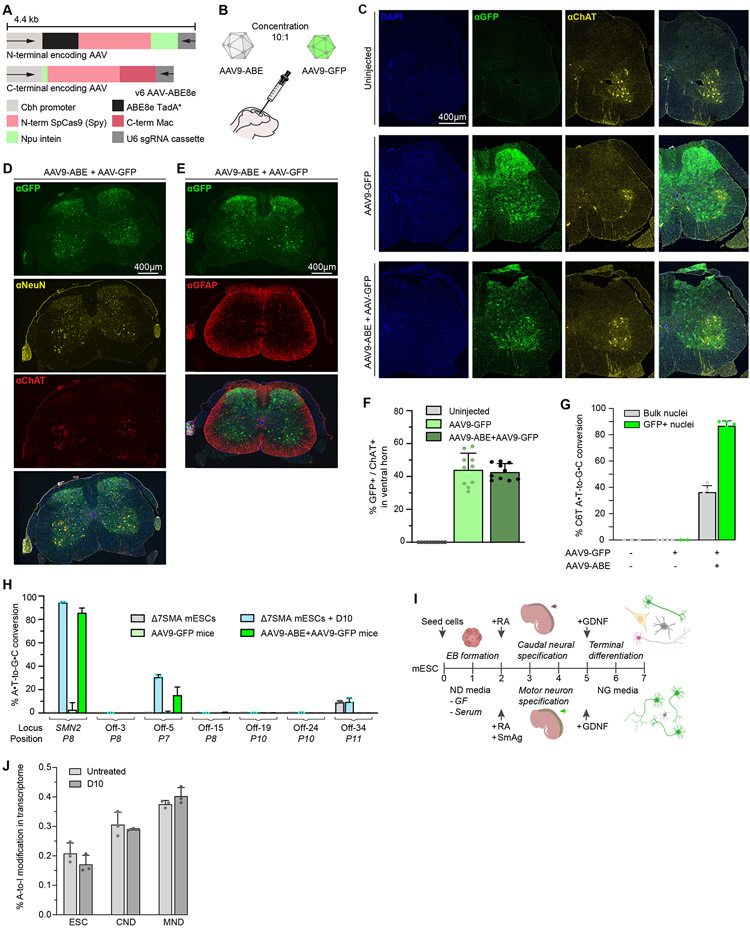
Fig. 3. Adenine base editing in Δ7SMA mice.
(A) Dual-AAV vectors encoding split-intein ABE8e-SpyMac and P8 sgRNA cassettes, v6 AAV9-ABE8e. (B) Neonatal ICV injections in Δ7SMA mice with AAV9-ABE, and AAV9-GFP as a transduction control. (C-E) Immunofluorescence images of lumbar spinal cord sections from wild-type Δ7SMA mice at 25 weeks old, ICV injected on PND0-1 with AAV9-ABE, AAV9-GFP, or uninjected as indicated. GFP indicates transduction, ChAT labels spinal motor neurons in the ventral horn, NeuN labels post-mitotic neurons, GFAP labels astrocytes, DAPI stains all nuclei. (F) Quantification of GFP and ChAT double-positive cells within the ventral horn (n=3). (G) Base editing in bulk and GFP+ flow-sorted nuclei of Δ7SMA mice treated with AAV9-ABE+AAV9-GFP (n=5), AAV9-GFP (n=4), or uninjected (n=3). (H) On-target and off-target editing following VIVO analysis of strategy D10 in Δ7SMA mESCs compared to AAV9-ABE+AAV9-GFP treatment in Δ7SMA mice. Bars show editing of the most frequently edited nucleotide at each locus, with the P# position shown in parenthesis. (I) Schematic of motor neuron differentiation (MND) and caudal-neural differentiation (CND) of Δ7SMA mESCs. (J) Whole transcriptome A-to-I RNA off-target editing analysis in Δ7SMAmESCs (n=3), and CND (n=3) and MND (n=3) differentiated cells stably expressing the D10 strategy.