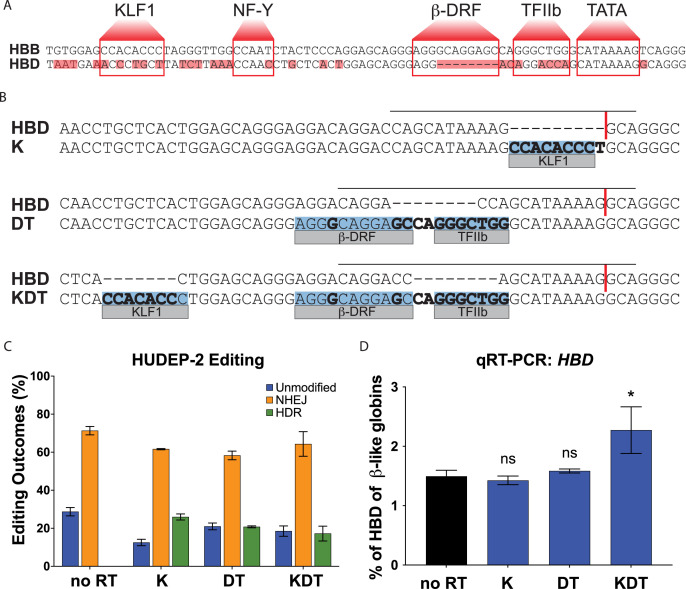
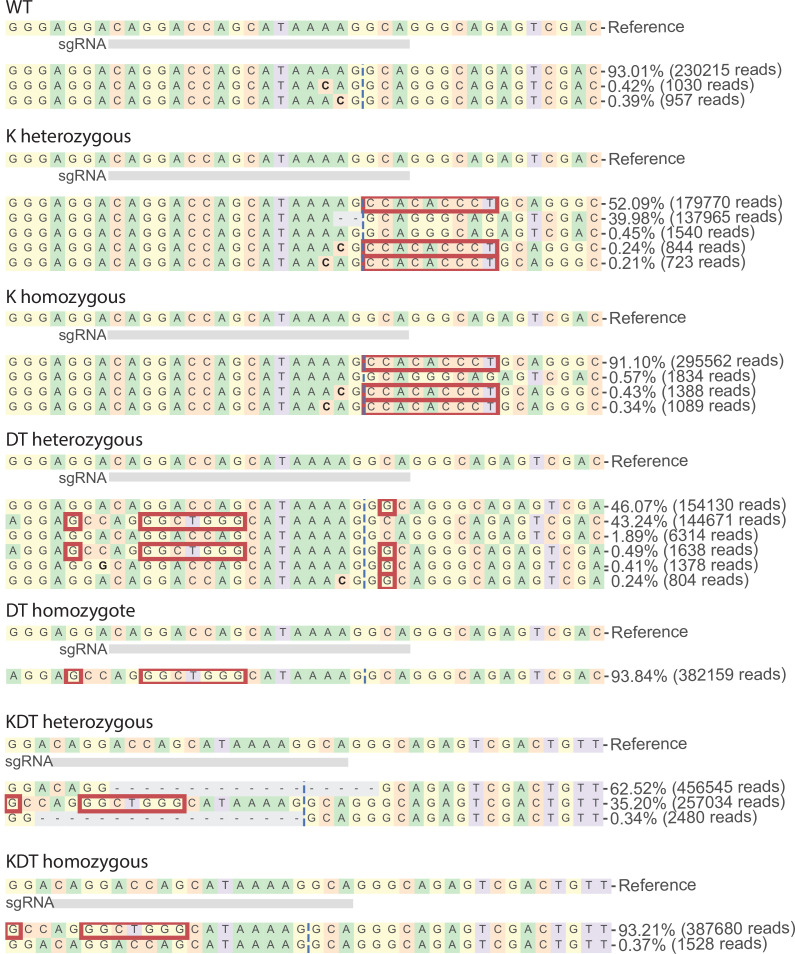
Figure 1. Targeting and design of the endogenous HBD promoter.
(A) Alignment of the HBB and HBD promoter sequences. Transcription factor binding sequences for KLF1, NF-Y, β-DRF, TFIIb, and TATA are shown in boxes and base pair mismatches between HBB and HBD are highlighted in red. (B) The repair template designs for insertions of KLF1 (K), β-DRF and TFIIb (DT), and KLF1, β-DRF, and TFIIb (KDT) directly compared to the HBD promoter. The inserted transcription factor binding sequences are highlighted in blue. Any base pair changes are in bold. The gRNA is indicated by a black horizontal line and the cut site is indicated by a red vertical line. (C) HUDEP-2 editing efficiencies showing percentages of unmodified, NHEJ, or HDR alleles. Conditions tested were Cas9 and sgRNA RNP with no repair template (no RT), K, DT, and KDT repair templates. This experiment was performed three times and the data is presented as mean ± SD of three biological replicates. (D) qRT-PCR of HBD after pooled editing of HUDEP-2 cells with no RT, K, DT, and KDT and 5 days of differentiation. Data is plotted as % of all β-like globins (HBB, HBG1/2, HBD). The three biological replicates from the editing experiment in (C) were each differentiated and the data is presented as mean ± SD of three biological replicates. p Value indicates paired, two-tailed student t test (ns, non-significant; *, p≤0.05; **, p≤0.01).