Fig. 2: AvPAL* deep mutational scanning (DMS) outcomes.
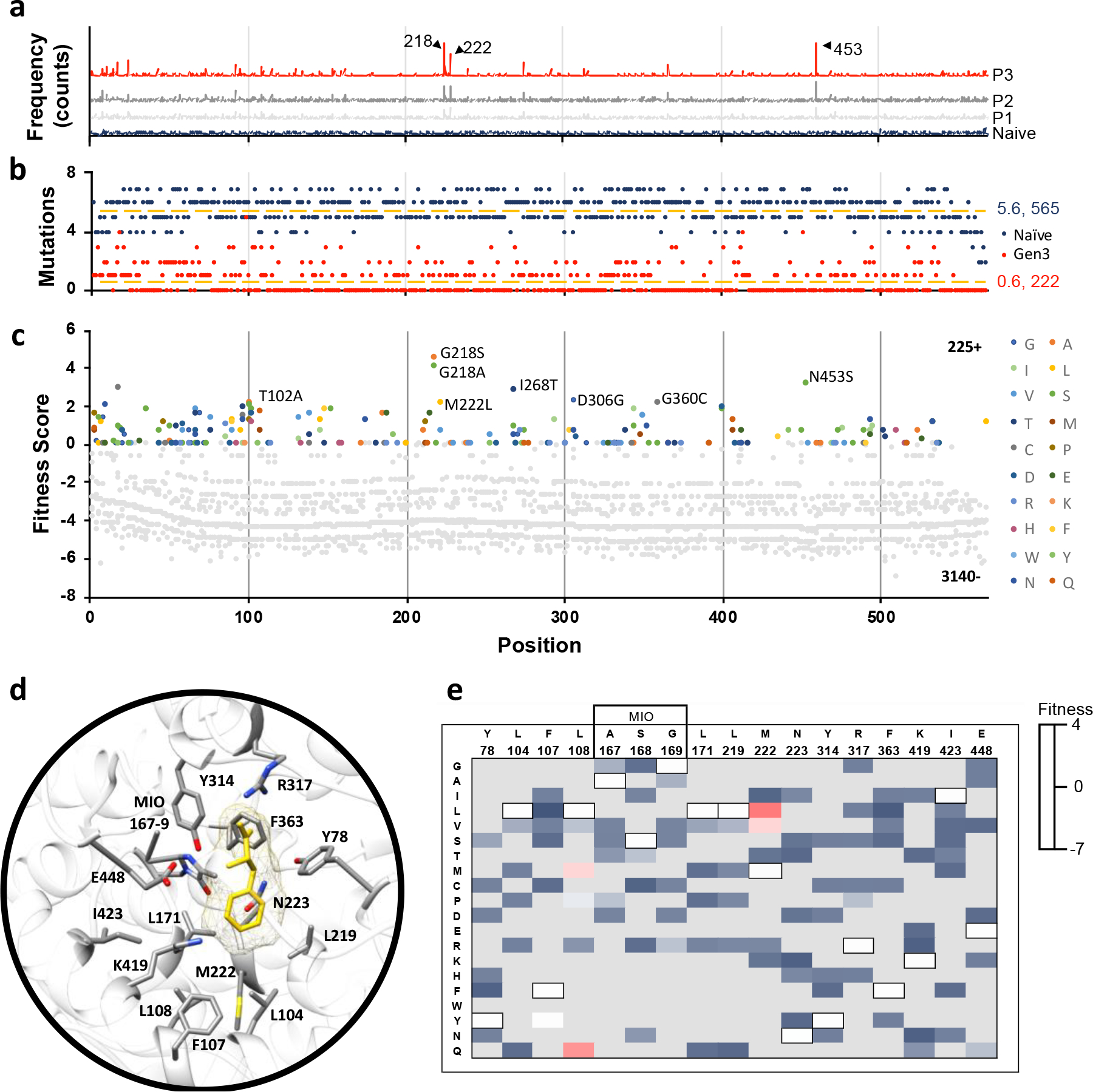
a) Mutation frequency in naïve and passages #1–3. Positions (a.a.) corresponding to the three most highly enriched positions are labeled (218, 222, and 453). b) Number of mutations sampled at each position in the naïve (dark blue) and passage #3 (red) libraries. The naïve library had an average of 5.6 mutations per position across all 565 positions. Passage #3 library had an average of 0.6 mutations per position across 222 positions. c) Fitness of all mutations present at all positions in the passage #3 library. Negative fitness denotes mutations that decrease in frequency over passages; the highest fitness mutations are labeled (G218A, G218S, M222L, I268T, D306G, G360C, and N453S). Position labels (x-axis) are the same for panels (a-c). From here we identified 79 functionally relevant sites of these, 7 positions T102, G218, M222L, I268, D306, G360 and N453 showed maximum fitness gradient and are thus referred to as hotspots. d) Active site residues of AvPAL* (grey sticks) with phenylalanine ligand (yellow) docked. e) Fitness heatmap of active site residues at passage #3. Wildtype residues of AvPAL* are bordered in black and listed above the residue position. Grey boxes indicate mutations not sampled in library.
