Fig. 3: Identification and location of highest fitness positions.
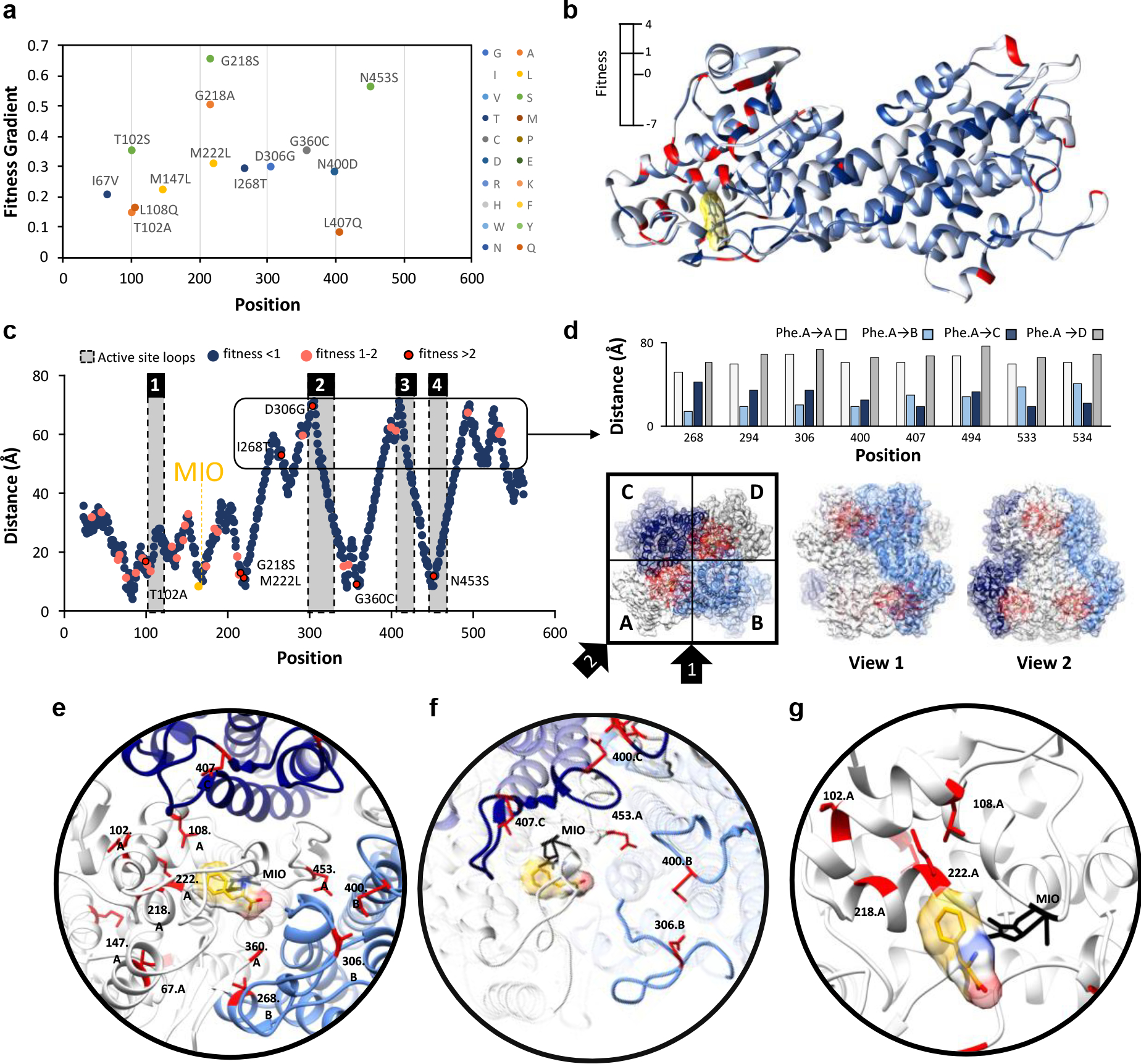
a) Gradient of fitness is calculated from passages 1–3. Only mutations with a frequency greater than zero in all passages and a passage #3 frequency greater than 0.625 % are shown. A positive gradient indicates increasing fitness across passages. b) Fitness score for the top variant found at that position is mapped to the structure of a single chain of AvPAL*. c) Distance of the α-carbon of docked phenylalanine to the α-carbon of every residue in AvPAL* within the same chain. Residues with a passage #3 fitness < 1 are dark blue, 1–2 are pink, and > 2 are red outlined in black. Active site loops are numbered 1–4 and shaded grey. d) High fitness residues that are distal from the active site are proximal to active sites of other subunits when visualized as part of a homotetramer. Chains A (white), B (light blue), C (dark blue), and D (grey) are shown from top and two side views. Residues near the active site are red. e) Locations of most fit residues relative to the active site of Chain A. Chains A (white), B (light blue), C (dark blue), and D (grey) are shown. The MIO adduct is black and the phenylalanine in yellow with red and blue oxygen and nitrogen atoms, respectively. Fit residues are colored red with sidechains shown. f) Residues 400, 407, 306, and 453 are in the active site loops (rendered opaque) previously identified as important for active site stability. g) Residues 102, 108, 218, and 222, are part of α-helices that form a surface within the active site, near the phenyl ring of the docked phenylalanine.
