Figure 4.
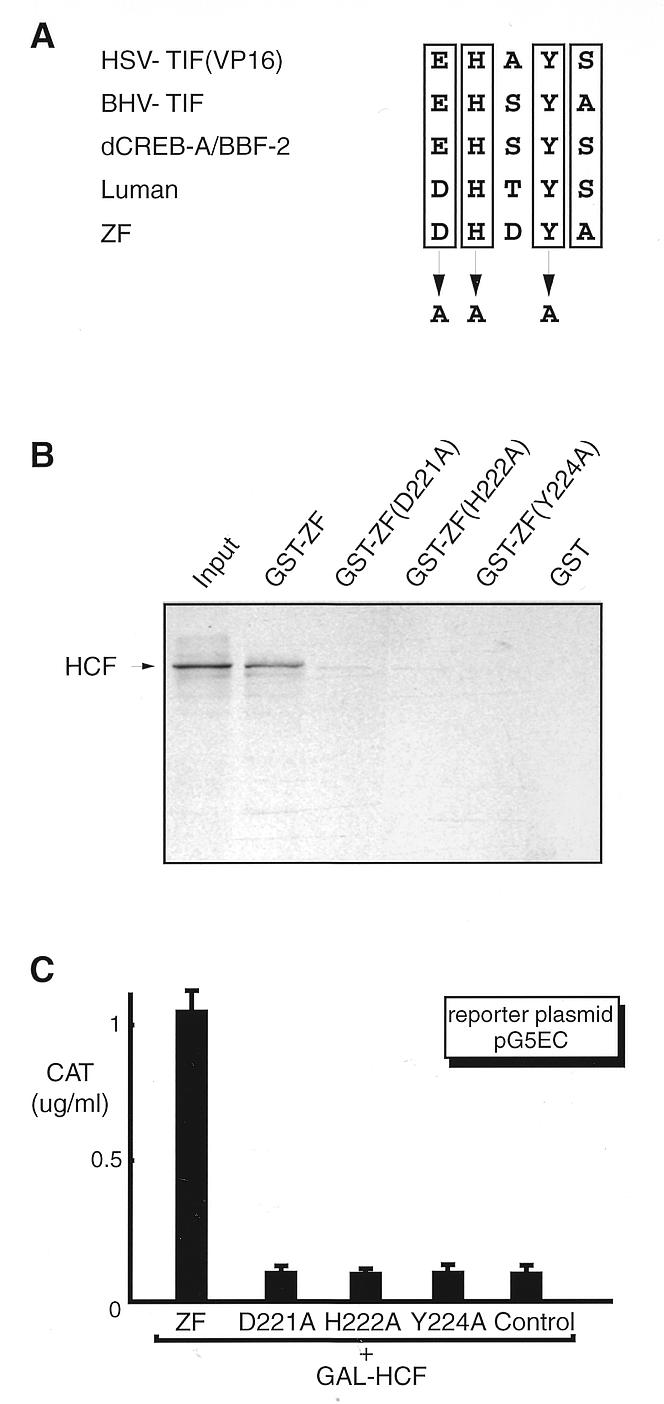
Mutagenesis study of the HCF-binding motif in ZF. (A) The consensus sequence, [D/E]HXY[S/A], found in VP16 and Luman as well as their homologs is proved to be a HCF-binding motif. To test whether this motif in ZF is also involved in HCF binding, we mutated two absolutely conserved residues, H222 and Y224, and the negatively charged residue D221 by site-directed mutagenesis. (B) GST pull-down assay. GST and all GST fusion proteins were produced in E.coli strain BL21(DE3), coupled to glutathione–Sepharose beads. After incubation with [35S]HCF, the beads were washed and analyzed by 10% SDS–PAGE. The lane labeled input represents one-tenth of the [35S]HCF incubation mixture. (C) Mammalian two-hybrid assay. Plasmids pcZF, pcZF(D221A), pcZF(H222A) and pcZF(Y224A) were introduced into COS7 cells separately with reporter pG5EC and the plasmid expressing GAL–HCF. An aliquot of 0.5 µg DNA was used for each plasmid. Plasmid pcDNA3 was used as a control. The CAT activity was measured by ELISA 48 h post-transfection.
