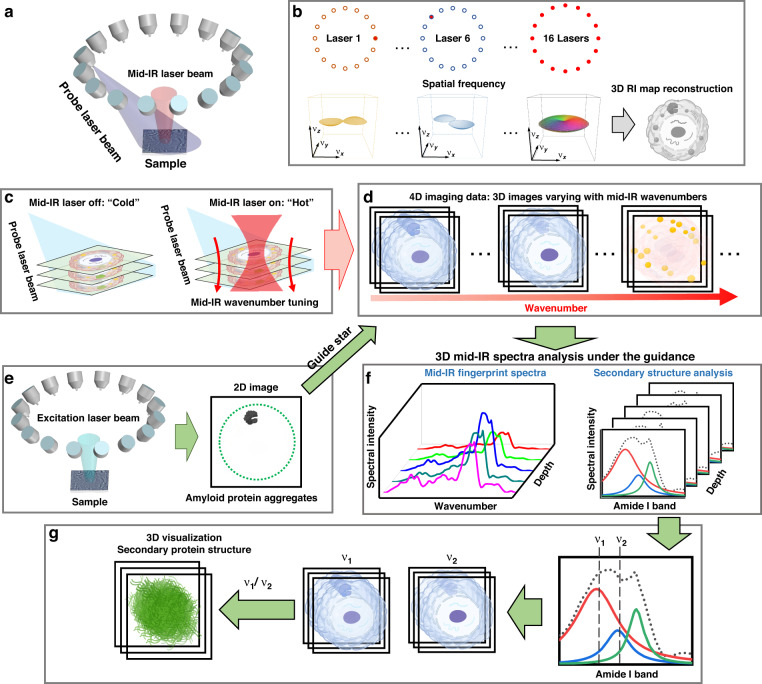
Fig. 1. FBS-IDT principle and workflow.
a Pump-probe 3D chemical imaging scheme. Each oblique pulsed probe beam (~450 nm) from a ring laser array illuminates the sample sequentially. A loosely focused pulsed mid-IR laser pump beam heats the sample periodically. b 3D RI map reconstruction scheme. Intensity imaging data from each probe beam illumination are mapped into the frequency domain. 3D RI map can be reconstructed by the inverse Fourier transform of the synthesized frequency domain information from all 16 probe beam detections. c “Cold” state: imaging without mid-IR pump beam illumination; “Hot” state: imaging with mid-IR pump beam illumination. d 4D hyperspectral chemical imaging. 3D chemical maps under different mid-IR wavenumbers are obtained in two steps: (1) subtracting “Hot” 3D RI maps from the “Cold” 3D RI maps under a particular wavenumber; (2) tuning the wavenumber to obtain 3D maps for different chemical compounds of interest. e Single-photon 2D fluorescence intensity imaging. To obtain the 2D guide star, both probe and pump beams are turned off while an excitation laser beam illuminates the sample. The 2D fluorescence images highlight the boundary of the amyloid protein aggregates. f Depth-resolved mid-IR fingerprint spectra generation and related protein secondary structure spectroscopic analysis. The depth-resolved mid-IR spectra are extracted from the 4D hyperspectral chemical imaging data under the guidance of the 2D image in (e). Secondary structures are analyzed by the deconvolution of the mid-IR amide I band. g 3D visualization of protein secondary structure for the amyloid protein aggregates. Based on the spectroscopic analysis results in (f), spectral positions for specific secondary protein structures are selected. 3D visualization of the secondary protein structures is obtained by extracting the mid-IR spectral ratio map between two 3D chemical images. Cell and protein aggregate icons in (b–e) and (g) are created and adapted from ref. 89